Visualizing water molecules in transmembrane proteins using radiolytic labeling methods
- PMID: 20047303
- PMCID: PMC2819031
- DOI: 10.1021/bi901889t
Visualizing water molecules in transmembrane proteins using radiolytic labeling methods
Abstract
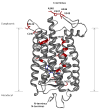
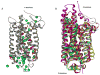
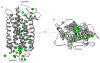
Essential to cells and their organelles, water is both shuttled to where it is needed and trapped within cellular compartments and structures. Moreover, ordered waters within protein structures often colocalize with strategically placed polar or charged groups critical for protein function, yet it is unclear if these ordered water molecules provide structural stabilization, mediate conformational changes in signaling, neutralize charged residues, or carry out a combination of all these functions. Structures of many integral membrane proteins, including G protein-coupled receptors (GPCRs), reveal the presence of ordered water molecules that may act like prosthetic groups in a manner quite unlike bulk water. Identification of "ordered" waters within a crystalline protein structure requires sufficient occupancy of water to enable its detection in the protein's X-ray diffraction pattern, and thus, the observed waters likely represent a subset of tightly bound functional waters. In this review, we highlight recent studies that suggest the structures of ordered waters within GPCRs are as conserved (and thus as important) as conserved side chains. In addition, methods of radiolysis, coupled to structural mass spectrometry (protein footprinting), reveal dynamic changes in water structure that mediate transmembrane signaling. The idea of water as a prosthetic group mediating chemical reaction dynamics is not new in fields such as catalysis. However, the concept of water as a mediator of conformational dynamics in signaling is just emerging, because of advances in both crystallographic structure determination and new methods of protein footprinting. Although oil and water do not mix, understanding the roles of water is essential to understanding the function of membrane proteins.
Figures

Similar articles
-
Detection of structural waters and their role in structural dynamics of rhodopsin activation.Methods Mol Biol. 2015;1271:97-111. doi: 10.1007/978-1-4939-2330-4_7. Methods Mol Biol. 2015. PMID: 25697519
-
Structural waters define a functional channel mediating activation of the GPCR, rhodopsin.Proc Natl Acad Sci U S A. 2009 Aug 25;106(34):14367-72. doi: 10.1073/pnas.0901074106. Epub 2009 Aug 13. Proc Natl Acad Sci U S A. 2009. PMID: 19706523 Free PMC article.
-
Role of bulk water in hydrolysis of the rhodopsin chromophore.J Biol Chem. 2011 May 27;286(21):18930-7. doi: 10.1074/jbc.M111.234583. Epub 2011 Apr 1. J Biol Chem. 2011. PMID: 21460218 Free PMC article.
-
Oxidative footprinting in the study of structure and function of membrane proteins: current state and perspectives.Biochem Soc Trans. 2015 Oct;43(5):983-94. doi: 10.1042/BST20150130. Biochem Soc Trans. 2015. PMID: 26517913 Review.
-
Using X-ray Footprinting and Mass Spectrometry to Study the Structure and Function of Membrane Proteins.Protein Pept Lett. 2019;26(1):44-54. doi: 10.2174/0929866526666181128142401. Protein Pept Lett. 2019. PMID: 30484402 Free PMC article. Review.
Cited by
-
Fast Photochemical Oxidation of Proteins Maps the Topology of Intrinsic Membrane Proteins: Light-Harvesting Complex 2 in a Nanodisc.Anal Chem. 2016 Sep 6;88(17):8827-34. doi: 10.1021/acs.analchem.6b01945. Epub 2016 Aug 16. Anal Chem. 2016. PMID: 27500903 Free PMC article.
-
Thermal properties of rhodopsin: insight into the molecular mechanism of dim-light vision.J Biol Chem. 2011 Aug 5;286(31):27622-9. doi: 10.1074/jbc.M111.233312. Epub 2011 Jun 9. J Biol Chem. 2011. PMID: 21659526 Free PMC article.
-
Chemical kinetic analysis of thermal decay of rhodopsin reveals unusual energetics of thermal isomerization and hydrolysis of Schiff base.J Biol Chem. 2011 Nov 4;286(44):38408-38416. doi: 10.1074/jbc.M111.280602. Epub 2011 Sep 15. J Biol Chem. 2011. PMID: 21921035 Free PMC article.
-
Use of protein cross-linking and radiolytic footprinting to elucidate PsbP and PsbQ interactions within higher plant Photosystem II.Proc Natl Acad Sci U S A. 2014 Nov 11;111(45):16178-83. doi: 10.1073/pnas.1415165111. Epub 2014 Oct 27. Proc Natl Acad Sci U S A. 2014. PMID: 25349426 Free PMC article.
-
A short story on how chromophore is hydrolyzed from rhodopsin for recycling.Bioessays. 2023 Sep;45(9):e2300068. doi: 10.1002/bies.202300068. Epub 2023 Jul 16. Bioessays. 2023. PMID: 37454357 Free PMC article. Review.
References
-
- Papoian GA, Ulander J, Wolynes PG. Role of water mediated interactions in protein-protein recognition landscapes. J Am Chem Soc. 2003;125:9170–9178. - PubMed
-
- Tame JR, Murshudov GN, Dodson EJ, Neil TK, Dodson GG, Higgins CF, Wilkinson AJ. The structural basis of sequence-independent peptide binding by OppA protein. Science. 1994;264:1578–1581. - PubMed
-
- Head-Gordon T, Brown S. Minimalist models for protein folding and design. Curr Opin Struct Biol. 2003;13:160–167. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P30 EB009998-01/EB/NIBIB NIH HHS/United States
- EY018085/EY/NEI NIH HHS/United States
- GM079191/GM/NIGMS NIH HHS/United States
- R01 EY009339-20/EY/NEI NIH HHS/United States
- R01 EB009688/EB/NIBIB NIH HHS/United States
- P30 EB009998/EB/NIBIB NIH HHS/United States
- EY009339/EY/NEI NIH HHS/United States
- EB09688/EB/NIBIB NIH HHS/United States
- R00 EY018085/EY/NEI NIH HHS/United States
- K99 EY018085/EY/NEI NIH HHS/United States
- P41 EB001979-26/EB/NIBIB NIH HHS/United States
- R01 EY009339/EY/NEI NIH HHS/United States
- R01 GM079191/GM/NIGMS NIH HHS/United States
- EB09998/EB/NIBIB NIH HHS/United States
- R01 GM079191-03/GM/NIGMS NIH HHS/United States
- P41 EB001979/EB/NIBIB NIH HHS/United States
LinkOut - more resources
Full Text Sources

