PLAZA: a comparative genomics resource to study gene and genome evolution in plants
- PMID: 20040540
- PMCID: PMC2814516
- DOI: 10.1105/tpc.109.071506
PLAZA: a comparative genomics resource to study gene and genome evolution in plants
Abstract
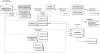
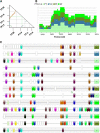
The number of sequenced genomes of representatives within the green lineage is rapidly increasing. Consequently, comparative sequence analysis has significantly altered our view on the complexity of genome organization, gene function, and regulatory pathways. To explore all this genome information, a centralized infrastructure is required where all data generated by different sequencing initiatives is integrated and combined with advanced methods for data mining. Here, we describe PLAZA, an online platform for plant comparative genomics (http://bioinformatics.psb.ugent.be/plaza/). This resource integrates structural and functional annotation of published plant genomes together with a large set of interactive tools to study gene function and gene and genome evolution. Precomputed data sets cover homologous gene families, multiple sequence alignments, phylogenetic trees, intraspecies whole-genome dot plots, and genomic colinearity between species. Through the integration of high confidence Gene Ontology annotations and tree-based orthology between related species, thousands of genes lacking any functional description are functionally annotated. Advanced query systems, as well as multiple interactive visualization tools, are available through a user-friendly and intuitive Web interface. In addition, detailed documentation and tutorials introduce the different tools, while the workbench provides an efficient means to analyze user-defined gene sets through PLAZA's interface. In conclusion, PLAZA provides a comprehensible and up-to-date research environment to aid researchers in the exploration of genome information within the green plant lineage.
Figures


Similar articles
-
A Guide to the PLAZA 3.0 Plant Comparative Genomic Database.Methods Mol Biol. 2017;1533:183-200. doi: 10.1007/978-1-4939-6658-5_10. Methods Mol Biol. 2017. PMID: 27987171
-
PLAZA 3.0: an access point for plant comparative genomics.Nucleic Acids Res. 2015 Jan;43(Database issue):D974-81. doi: 10.1093/nar/gku986. Epub 2014 Oct 16. Nucleic Acids Res. 2015. PMID: 25324309 Free PMC article.
-
PLAZA 5.0: extending the scope and power of comparative and functional genomics in plants.Nucleic Acids Res. 2022 Jan 7;50(D1):D1468-D1474. doi: 10.1093/nar/gkab1024. Nucleic Acids Res. 2022. PMID: 34747486 Free PMC article.
-
An Experimental Approach to Genome Annotation: This report is based on a colloquium sponsored by the American Academy of Microbiology held July 19-20, 2004, in Washington, DC.Washington (DC): American Society for Microbiology; 2004. Washington (DC): American Society for Microbiology; 2004. PMID: 33001599 Free Books & Documents. Review.
-
Mitochondrial Disease Sequence Data Resource (MSeqDR): a global grass-roots consortium to facilitate deposition, curation, annotation, and integrated analysis of genomic data for the mitochondrial disease clinical and research communities.Mol Genet Metab. 2015 Mar;114(3):388-96. doi: 10.1016/j.ymgme.2014.11.016. Epub 2014 Dec 4. Mol Genet Metab. 2015. PMID: 25542617 Free PMC article. Review.
Cited by
-
The emergence of molecular profiling and omics techniques in seagrass biology; furthering our understanding of seagrasses.Funct Integr Genomics. 2016 Sep;16(5):465-80. doi: 10.1007/s10142-016-0501-4. Epub 2016 Jul 21. Funct Integr Genomics. 2016. PMID: 27443314 Review.
-
The hybrid four-CBS-domain KINβγ subunit functions as the canonical γ subunit of the plant energy sensor SnRK1.Plant J. 2013 Jul;75(1):11-25. doi: 10.1111/tpj.12192. Epub 2013 May 15. Plant J. 2013. PMID: 23551663 Free PMC article.
-
Plant F-box protein evolution is determined by lineage-specific timing of major gene family expansion waves.PLoS One. 2013 Jul 19;8(7):e68672. doi: 10.1371/journal.pone.0068672. Print 2013. PLoS One. 2013. PMID: 23904908 Free PMC article.
-
Transcriptome analysis indicates considerable divergence in alternative splicing between duplicated genes in Arabidopsis thaliana.Genetics. 2014 Dec;198(4):1473-81. doi: 10.1534/genetics.114.169466. Epub 2014 Oct 16. Genetics. 2014. PMID: 25326238 Free PMC article.
-
Integrative Approaches to Enhance Understanding of Plant Metabolic Pathway Structure and Regulation.Plant Physiol. 2015 Nov;169(3):1499-511. doi: 10.1104/pp.15.01006. Epub 2015 Sep 14. Plant Physiol. 2015. PMID: 26371234 Free PMC article.
References
-
- Allen, B.S., Stein, J.L., Stein, G.S., and Ostrer, H. (1991). Single-copy flanking sequences in human histone gene clusters map to chromosomes 1 and 6. Genomics 10 486–488. - PubMed
-
- Arabidopsis Genome Initiative (2000). Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408 796–815. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

