CD36 ligands promote sterile inflammation through assembly of a Toll-like receptor 4 and 6 heterodimer
- PMID: 20037584
- PMCID: PMC2809046
- DOI: 10.1038/ni.1836
CD36 ligands promote sterile inflammation through assembly of a Toll-like receptor 4 and 6 heterodimer
Abstract
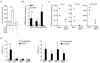
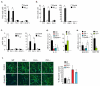
In atherosclerosis and Alzheimer's disease, deposition of the altered self components oxidized low-density lipoprotein (LDL) and amyloid-beta triggers a protracted sterile inflammatory response. Although chronic stimulation of the innate immune system is believed to underlie the pathology of these diseases, the molecular mechanisms of activation remain unclear. Here we show that oxidized LDL and amyloid-beta trigger inflammatory signaling through a heterodimer of Toll-like receptors 4 and 6. Assembly of this newly identified heterodimer is regulated by signals from the scavenger receptor CD36, a common receptor for these disparate ligands. Our results identify CD36-TLR4-TLR6 activation as a common molecular mechanism by which atherogenic lipids and amyloid-beta stimulate sterile inflammation and suggest a new model of TLR heterodimerization triggered by coreceptor signaling events.
Figures
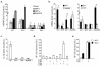
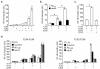


Similar articles
-
Intramembrane attenuation of the TLR4-TLR6 dimer impairs receptor assembly and reduces microglia-mediated neurodegeneration.J Biol Chem. 2017 Aug 11;292(32):13415-13427. doi: 10.1074/jbc.M117.784983. Epub 2017 Jun 27. J Biol Chem. 2017. PMID: 28655763 Free PMC article.
-
CD36 coordinates NLRP3 inflammasome activation by facilitating intracellular nucleation of soluble ligands into particulate ligands in sterile inflammation.Nat Immunol. 2013 Aug;14(8):812-20. doi: 10.1038/ni.2639. Epub 2013 Jun 30. Nat Immunol. 2013. PMID: 23812099 Free PMC article.
-
Cloning and expression of an anti-LDL(-) single-chain variable fragment, and its inhibitory effect on experimental atherosclerosis.MAbs. 2013 Sep-Oct;5(5):763-75. doi: 10.4161/mabs.25859. Epub 2013 Jul 25. MAbs. 2013. PMID: 23924793 Free PMC article.
-
Saturated fatty acids trigger TLR4-mediated inflammatory response.Atherosclerosis. 2016 Jan;244:211-5. doi: 10.1016/j.atherosclerosis.2015.11.015. Epub 2015 Dec 2. Atherosclerosis. 2016. PMID: 26687466 Review.
-
CD36 in Atherosclerosis: Pathophysiological Mechanisms and Therapeutic Implications.Curr Atheroscler Rep. 2020 Aug 9;22(10):59. doi: 10.1007/s11883-020-00870-8. Curr Atheroscler Rep. 2020. PMID: 32772254 Review.
Cited by
-
Increased serum levels of advanced glycation end products due to induced molting in hen layers trigger a proinflammatory response by peripheral blood leukocytes.Poult Sci. 2020 Jul;99(7):3452-3462. doi: 10.1016/j.psj.2020.04.009. Epub 2020 Apr 26. Poult Sci. 2020. PMID: 32616239 Free PMC article.
-
Innate immunity and neuroinflammation.Mediators Inflamm. 2013;2013:342931. doi: 10.1155/2013/342931. Epub 2013 Jun 15. Mediators Inflamm. 2013. PMID: 23843682 Free PMC article. Review.
-
Malondialdehyde Epitopes as Targets of Immunity and the Implications for Atherosclerosis.Adv Immunol. 2016;131:1-59. doi: 10.1016/bs.ai.2016.02.001. Epub 2016 Apr 5. Adv Immunol. 2016. PMID: 27235680 Free PMC article. Review.
-
CD36 modulates proinflammatory cytokine responses to Plasmodium falciparum glycosylphosphatidylinositols and merozoites by dendritic cells.Parasite Immunol. 2012 Jul;34(7):372-82. doi: 10.1111/j.1365-3024.2012.01367.x. Parasite Immunol. 2012. PMID: 22486596 Free PMC article.
-
Phenotype and Response to PAMPs of Human Monocyte-Derived Foam Cells Obtained by Long-Term Culture in the Presence of oxLDLs.Front Immunol. 2020 Aug 4;11:1592. doi: 10.3389/fimmu.2020.01592. eCollection 2020. Front Immunol. 2020. PMID: 32849539 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

