Epstein-Barr virus DNase (BGLF5) induces genomic instability in human epithelial cells
- PMID: 20034954
- PMCID: PMC2847232
- DOI: 10.1093/nar/gkp1169
Epstein-Barr virus DNase (BGLF5) induces genomic instability in human epithelial cells
Abstract
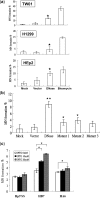
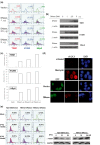
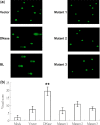
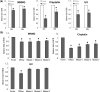
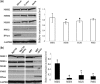
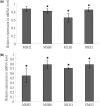
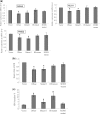
Epstein-Barr Virus (EBV) DNase (BGLF5) is an alkaline nuclease and has been suggested to be important in the viral life cycle. However, its effect on host cells remains unknown. Serological and histopathological studies implied that EBV DNase seems to be correlated with carcinogenesis. Therefore, we investigate the effect of EBV DNase on epithelial cells. Here, we report that expression of EBV DNase induces increased formation of micronucleus, an indicator of genomic instability, in human epithelial cells. We also demonstrate, using gammaH2AX formation and comet assay, that EBV DNase induces DNA damage. Furthermore, using host cell reactivation assay, we find that EBV DNase expression repressed damaged DNA repair in various epithelial cells. Western blot and quantitative PCR analyses reveal that expression of repair-related genes is reduced significantly in cells expressing EBV DNase. Host shut-off mutants eliminate shut-off expression of repair genes and repress damaged DNA repair, suggesting that shut-off function of BGLF5 contributes to repression of DNA repair. In addition, EBV DNase caused chromosomal aberrations and increased the microsatellite instability (MSI) and frequency of genetic mutation in human epithelial cells. Together, we propose that EBV DNase induces genomic instability in epithelial cells, which may be through induction of DNA damage and also repression of DNA repair, subsequently increases MSI and genetic mutations, and may contribute consequently to the carcinogenesis of human epithelial cells.
Figures
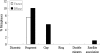
Similar articles
-
Epstein-Barr virus latent membrane protein 1 induces micronucleus formation, represses DNA repair and enhances sensitivity to DNA-damaging agents in human epithelial cells.Oncogene. 2004 Apr 1;23(14):2531-9. doi: 10.1038/sj.onc.1207375. Oncogene. 2004. PMID: 14716302
-
Screening and identification of emodin as an EBV DNase inhibitor to prevent its biological functions.Virol J. 2023 Jul 13;20(1):148. doi: 10.1186/s12985-023-02107-x. Virol J. 2023. PMID: 37443068 Free PMC article.
-
The "Bridge" in the Epstein-Barr virus alkaline exonuclease protein BGLF5 contributes to shutoff activity during productive infection.J Virol. 2012 Sep;86(17):9175-87. doi: 10.1128/JVI.00309-12. Epub 2012 Jun 13. J Virol. 2012. PMID: 22696660 Free PMC article.
-
Epstein-Barr Virus Hijacks DNA Damage Response Transducers to Orchestrate Its Life Cycle.Viruses. 2017 Nov 16;9(11):341. doi: 10.3390/v9110341. Viruses. 2017. PMID: 29144413 Free PMC article. Review.
-
Mechanisms of formation of chromosomal aberrations: insights from studies with DNA repair-deficient cells.Cytogenet Genome Res. 2004;104(1-4):95-9. doi: 10.1159/000077471. Cytogenet Genome Res. 2004. PMID: 15162020 Review.
Cited by
-
The Immunomodulatory Capacity of an Epstein-Barr Virus Abortive Lytic Cycle: Potential Contribution to Viral Tumorigenesis.Cancers (Basel). 2018 Mar 30;10(4):98. doi: 10.3390/cancers10040098. Cancers (Basel). 2018. PMID: 29601503 Free PMC article. Review.
-
Contribution of Epstein-Barr Virus Lytic Proteins to Cancer Hallmarks and Implications from Other Oncoviruses.Cancers (Basel). 2023 Apr 2;15(7):2120. doi: 10.3390/cancers15072120. Cancers (Basel). 2023. PMID: 37046781 Free PMC article. Review.
-
Viral-Targeted Strategies Against EBV-Associated Lymphoproliferative Diseases.Front Oncol. 2019 Feb 26;9:81. doi: 10.3389/fonc.2019.00081. eCollection 2019. Front Oncol. 2019. PMID: 30873380 Free PMC article. Review.
-
The Application of Emodin Treatment on Nasopharyngeal Carcinoma Therapy.Biomedicines. 2024 Feb 21;12(3):486. doi: 10.3390/biomedicines12030486. Biomedicines. 2024. PMID: 38540100 Free PMC article. Review.
-
An Epstein-Barr virus protein interaction map reveals NLRP3 inflammasome evasion via MAVS UFMylation.Mol Cell. 2023 Jul 6;83(13):2367-2386.e15. doi: 10.1016/j.molcel.2023.05.018. Epub 2023 Jun 12. Mol Cell. 2023. PMID: 37311461 Free PMC article.

