Complex landscapes of somatic rearrangement in human breast cancer genomes
- PMID: 20033038
- PMCID: PMC3398135
- DOI: 10.1038/nature08645
Complex landscapes of somatic rearrangement in human breast cancer genomes
Abstract
Multiple somatic rearrangements are often found in cancer genomes; however, the underlying processes of rearrangement and their contribution to cancer development are poorly characterized. Here we use a paired-end sequencing strategy to identify somatic rearrangements in breast cancer genomes. There are more rearrangements in some breast cancers than previously appreciated. Rearrangements are more frequent over gene footprints and most are intrachromosomal. Multiple rearrangement architectures are present, but tandem duplications are particularly common in some cancers, perhaps reflecting a specific defect in DNA maintenance. Short overlapping sequences at most rearrangement junctions indicate that these have been mediated by non-homologous end-joining DNA repair, although varying sequence patterns indicate that multiple processes of this type are operative. Several expressed in-frame fusion genes were identified but none was recurrent. The study provides a new perspective on cancer genomes, highlighting the diversity of somatic rearrangements and their potential contribution to cancer development.
Figures
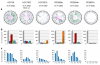

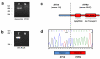
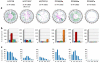
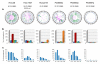


Similar articles
-
Architectures of somatic genomic rearrangement in human cancer amplicons at sequence-level resolution.Genome Res. 2007 Sep;17(9):1296-303. doi: 10.1101/gr.6522707. Epub 2007 Aug 3. Genome Res. 2007. PMID: 17675364 Free PMC article.
-
Somatic structural rearrangements in genetically engineered mouse mammary tumors.Genome Biol. 2010;11(10):R100. doi: 10.1186/gb-2010-11-10-r100. Epub 2010 Oct 13. Genome Biol. 2010. PMID: 20942901 Free PMC article.
-
Comprehensive long-span paired-end-tag mapping reveals characteristic patterns of structural variations in epithelial cancer genomes.Genome Res. 2011 May;21(5):665-75. doi: 10.1101/gr.113555.110. Epub 2011 Apr 5. Genome Res. 2011. PMID: 21467267 Free PMC article.
-
Mechanistic origins of diverse genome rearrangements in cancer.Semin Cell Dev Biol. 2022 Mar;123:100-109. doi: 10.1016/j.semcdb.2021.03.003. Epub 2021 Apr 3. Semin Cell Dev Biol. 2022. PMID: 33824062 Free PMC article. Review.
-
Processes shaping cancer genomes - From mitotic defects to chromosomal rearrangements.DNA Repair (Amst). 2021 Nov;107:103207. doi: 10.1016/j.dnarep.2021.103207. Epub 2021 Aug 10. DNA Repair (Amst). 2021. PMID: 34425515 Review.
Cited by
-
53BP1 deficiency leads to hyperrecombination using break-induced replication (BIR).Nat Commun. 2024 Oct 5;15(1):8648. doi: 10.1038/s41467-024-52916-z. Nat Commun. 2024. PMID: 39368985 Free PMC article.
-
53BP1 deficiency leads to hyperrecombination using break-induced replication (BIR).bioRxiv [Preprint]. 2024 Sep 13:2024.09.11.612483. doi: 10.1101/2024.09.11.612483. bioRxiv. 2024. Update in: Nat Commun. 2024 Oct 5;15(1):8648. doi: 10.1038/s41467-024-52916-z. PMID: 39314326 Free PMC article. Updated. Preprint.
-
Prognostic mutation signature would serve as a potential prognostic predictor in patients with diffuse large B-cell lymphoma.Sci Rep. 2024 Mar 14;14(1):6161. doi: 10.1038/s41598-024-56583-4. Sci Rep. 2024. PMID: 38485750 Free PMC article.
-
Intragenic Rearrangement Burden Associates with Immune Cell Infiltration and Response to Immune Checkpoint Blockade in Cancer.Cancer Immunol Res. 2024 Mar 4;12(3):287-295. doi: 10.1158/2326-6066.CIR-22-0637. Cancer Immunol Res. 2024. PMID: 38345376 Free PMC article.
-
Diffuse Gliomas with FGFR3-TACC3 Fusions: Oncogenic Mechanisms, Hallmarks, and Therapeutic Perspectives.Cancers (Basel). 2023 Nov 23;15(23):5555. doi: 10.3390/cancers15235555. Cancers (Basel). 2023. PMID: 38067258 Free PMC article. Review.
References
-
- Mitelman F, Johansson B, Mertens F. Fusion genes and rearranged genes as a linear function of chromosome aberrations in cancer. Nat Genet. 2004;36:331–4. - PubMed
-
- Mitelman F, Johansson B, Mertens F. The impact of translocations and gene fusions on cancer causation. Nat Rev Cancer. 2007;7:233–45. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

