Elongation in translation as a dynamic interaction among the ribosome, tRNA, and elongation factors EF-G and EF-Tu
- PMID: 20025795
- PMCID: PMC2832932
- DOI: 10.1017/S0033583509990060
Elongation in translation as a dynamic interaction among the ribosome, tRNA, and elongation factors EF-G and EF-Tu
Abstract
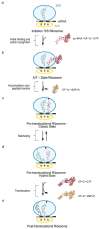
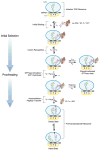
The ribosome is a complex macromolecular machine that translates the message encoded in the messenger RNA and synthesizes polypeptides by linking the individual amino acids carried by the cognate transfer RNAs (tRNAs). The protein elongation cycle, during which the tRNAs traverse the ribosome in a coordinated manner along a path of more than 100 A, is facilitated by large-scale rearrangements of the ribosome. These rearrangements go hand in hand with conformational changes of tRNA as well as elongation factors EF-Tu and EF-G - GTPases that catalyze tRNA delivery and translocation, respectively. This review focuses on the structural data related to the dynamics of the ribosomal machinery, which are the basis, in conjunction with existing biochemical, kinetic, and fluorescence resonance energy transfer data, of our knowledge of the decoding and translocation steps of protein elongation.
Figures
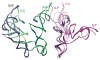

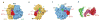


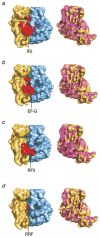
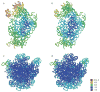
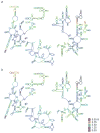

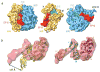


Similar articles
-
Molecular dynamics of ribosomal elongation factors G and Tu.Eur Biophys J. 2011 Mar;40(3):289-303. doi: 10.1007/s00249-010-0647-2. Epub 2010 Dec 9. Eur Biophys J. 2011. PMID: 21152913 Free PMC article.
-
Translocation as continuous movement through the ribosome.RNA Biol. 2016 Dec;13(12):1197-1203. doi: 10.1080/15476286.2016.1240140. Epub 2016 Nov 1. RNA Biol. 2016. PMID: 27801619 Free PMC article.
-
Elongation factors on the ribosome.Curr Opin Struct Biol. 2005 Jun;15(3):349-54. doi: 10.1016/j.sbi.2005.05.004. Curr Opin Struct Biol. 2005. PMID: 15922593 Review.
-
Highly conserved base A55 of 16S ribosomal RNA is important for the elongation cycle of protein synthesis.Biochemistry. 2013 Sep 24;52(38):6695-701. doi: 10.1021/bi4008879. Epub 2013 Sep 11. Biochemistry. 2013. PMID: 24025161
-
Ribosome Structure, Function, and Early Evolution.Int J Mol Sci. 2018 Dec 21;20(1):40. doi: 10.3390/ijms20010040. Int J Mol Sci. 2018. PMID: 30583477 Free PMC article. Review.
Cited by
-
Why nature really chose phosphate.Q Rev Biophys. 2013 Feb;46(1):1-132. doi: 10.1017/S0033583512000157. Epub 2013 Jan 15. Q Rev Biophys. 2013. PMID: 23318152 Free PMC article. Review.
-
Unraveling the dynamics of ribosome translocation.Curr Opin Struct Biol. 2012 Dec;22(6):804-14. doi: 10.1016/j.sbi.2012.09.004. Epub 2012 Nov 8. Curr Opin Struct Biol. 2012. PMID: 23142574 Free PMC article. Review.
-
Unveiling the inhibition mechanism of Clostridioides difficile by Bifidobacterium longum via multiomics approach.Front Microbiol. 2023 Nov 8;14:1293149. doi: 10.3389/fmicb.2023.1293149. eCollection 2023. Front Microbiol. 2023. PMID: 38029200 Free PMC article.
-
From DNA to proteins via the ribosome: structural insights into the workings of the translation machinery.Hum Genomics. 2010 Apr;4(4):226-37. doi: 10.1186/1479-7364-4-4-226. Hum Genomics. 2010. PMID: 20511136 Free PMC article. Review.
-
Heterogeneity of the translational machinery: Variations on a common theme.Biochimie. 2015 Jul;114:39-47. doi: 10.1016/j.biochi.2014.12.011. Epub 2014 Dec 24. Biochimie. 2015. PMID: 25542647 Free PMC article. Review.
References
-
- Abel KM, Yoder MD, Hilgenfeld R, Jurnak F. An α to β conformational switch in EF-Tu. Structure. 1996;4:1153–1159. - PubMed
-
- Agrawal RK, Heagle AB, Penczek P, Grassucci R, Frank J. EF-G-dependent GTP hydrolysis induces translocation accompanied by large conformational changes in the 70S ribosome. Nature Structural Biology. 1999;6:643–647. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

