Augmented latent membrane protein 1 expression from Epstein-Barr virus episomes with minimal terminal repeats
- PMID: 20015988
- PMCID: PMC2820940
- DOI: 10.1128/JVI.01972-09
Augmented latent membrane protein 1 expression from Epstein-Barr virus episomes with minimal terminal repeats
Abstract
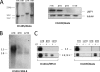
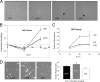
The major oncogene of the Epstein-Barr virus (EBV), latent membrane protein 1 (LMP1), can be expressed from either of two promoters, ED-L1 or L1-TR, producing mRNAs of 2.8 kb or 3.5 kb, respectively. L1-TR, active in nasopharyngeal carcinoma and Hodgkin's lymphoma, is located within the first of a highly variable reiteration of terminal repeat (TR) sequences that are joined by random recombination upon circularization of the linear genome at entry into cells. To determine whether the resultant TR number affects LMP1 promoter activity, we isolated single-cell clones bearing episomes of distinct TR numbers (6TR to 12TR) from epithelial cells newly infected with EBV. LMP1 mRNA levels correlated directly with the quantity of LMP1 protein expressed but varied inversely to TR number. Unexpectedly, the 3.5-kb transcript predominated only at lower TR reiterations. Diminished L1-TR activity in the context of a higher TR count was confirmed with a green fluorescent protein (GFP) reporter construct driven by L1-TR. Various levels of LMP1, expressed from virus isogenic in all but TR number, produced divergent morphological and growth phenotypes in each cell clone. Abundant LMP1 in 6TR cells yielded a relatively cytostatic state compared to the proliferative one produced by intermediate and smaller amounts in 8TR and 12TR clones. These findings suggest that the diversification of TR number, inherent in a round of EBV reactivation and reinfection, may itself be a component of the oncogenic process. The replicative burst preceding onset of many EBV-linked cancers may increase the likelihood that LMP1 levels compatible with clonal outgrowth are achieved in a subset of infected cells.
Figures



Similar articles
-
Linkage between STAT regulation and Epstein-Barr virus gene expression in tumors.J Virol. 2001 Mar;75(6):2929-37. doi: 10.1128/JVI.75.6.2929-2937.2001. J Virol. 2001. PMID: 11222718 Free PMC article.
-
Length of Epstein-Barr virus termini as a determinant of epithelial cell clonal emergence.J Virol. 2003 Aug;77(15):8555-61. doi: 10.1128/jvi.77.15.8555-8561.2003. J Virol. 2003. PMID: 12857925 Free PMC article.
-
Regulation of DNA Damage Signaling and Cell Death Responses by Epstein-Barr Virus Latent Membrane Protein 1 (LMP1) and LMP2A in Nasopharyngeal Carcinoma Cells.J Virol. 2015 Aug;89(15):7612-24. doi: 10.1128/JVI.00958-15. Epub 2015 May 13. J Virol. 2015. PMID: 25972552 Free PMC article.
-
LMP1-Induced Sumoylation Influences the Maintenance of Epstein-Barr Virus Latency through KAP1.J Virol. 2015 Aug;89(15):7465-77. doi: 10.1128/JVI.00711-15. Epub 2015 May 6. J Virol. 2015. PMID: 25948750 Free PMC article.
-
Differentiation-Dependent LMP1 Expression Is Required for Efficient Lytic Epstein-Barr Virus Reactivation in Epithelial Cells.J Virol. 2017 Mar 29;91(8):e02438-16. doi: 10.1128/JVI.02438-16. Print 2017 Apr 15. J Virol. 2017. PMID: 28179525 Free PMC article.
Cited by
-
Epigenetic deregulation of the LMP1/LMP2 locus of Epstein-Barr virus by mutation of a single CTCF-cohesin binding site.J Virol. 2014 Feb;88(3):1703-13. doi: 10.1128/JVI.02209-13. Epub 2013 Nov 20. J Virol. 2014. PMID: 24257606 Free PMC article.
-
TRAF3 signaling: Competitive binding and evolvability of adaptive viral molecular mimicry.Biochim Biophys Acta. 2016 Nov;1860(11 Pt B):2646-55. doi: 10.1016/j.bbagen.2016.05.021. Epub 2016 May 18. Biochim Biophys Acta. 2016. PMID: 27208423 Free PMC article. Review.
-
An atlas of the Epstein-Barr virus transcriptome and epigenome reveals host-virus regulatory interactions.Cell Host Microbe. 2012 Aug 16;12(2):233-45. doi: 10.1016/j.chom.2012.06.008. Cell Host Microbe. 2012. PMID: 22901543 Free PMC article.
-
Epstein-Barr virus-induced epigenetic alterations following transient infection.Int J Cancer. 2013 May 1;132(9):2076-86. doi: 10.1002/ijc.27893. Epub 2012 Nov 2. Int J Cancer. 2013. PMID: 23047626 Free PMC article.
-
EBV Association with Lymphomas and Carcinomas in the Oral Compartment.Viruses. 2022 Dec 1;14(12):2700. doi: 10.3390/v14122700. Viruses. 2022. PMID: 36560704 Free PMC article. Review.
References
-
- Baer, R. J., A. T. Bankier, M. D. Biggin, P. L. Deininger, P. J. Farrell, T. J. Gibson, G. F. Hatfill, G. S. Hudson, S. C. Satchwell, C. Seguin, P. S. Tuffnell, and B. C. Barrell. 1984. DNA sequences and expression of the B95-8 Epstein-Barr virus genome. Nature 310:207-211. - PubMed
-
- Blackwood, E. M., and J. T. Kadonaga. 1998. Going the distance: a current review of enhancer action. Science 281:60-63. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

