Molecular evolution of sex-biased genes in the Drosophila ananassae subgroup
- PMID: 20015359
- PMCID: PMC2809073
- DOI: 10.1186/1471-2148-9-291
Molecular evolution of sex-biased genes in the Drosophila ananassae subgroup
Abstract
Background: Genes with sex-biased expression often show rapid molecular evolution between species. Previous population genetic and comparative genomic studies of Drosophila melanogaster and D. simulans revealed that male-biased genes have especially high rates of adaptive evolution. To test if this is also the case for other lineages within the melanogaster group, we investigated gene expression in D. ananassae, a species that occurs in structured populations in tropical and subtropical regions. We used custom-made microarrays and published microarray data to characterize the sex-biased expression of 129 D. ananassae genes whose D. melanogaster orthologs had been classified previously as male-biased, female-biased, or unbiased in their expression and had been studied extensively at the population-genetic level. For 43 of these genes we surveyed DNA sequence polymorphism in a natural population of D. ananassae and determined divergence to the sister species D. atripex and D. phaeopleura.
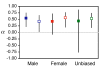
Results: Sex-biased expression is generally conserved between D. melanogaster and D. ananassae, with the majority of genes exhibiting the same bias in the two species. However, about one-third of the genes have either gained or lost sex-biased expression in one of the species and a small proportion of genes (approximately 4%) have changed bias from one sex to the other. The male-biased genes of D. ananassae show evidence of positive selection acting at the protein level. However, the signal of adaptive protein evolution for male-biased genes is not as strong in D. ananassae as it is in D. melanogaster and is limited to genes with conserved male-biased expression in both species. Within D. ananassae, a significant signal of adaptive evolution is also detected for female-biased and unbiased genes.
Conclusions: Our findings extend previous observations of widespread adaptive protein evolution to an independent Drosophila lineage, the D. ananassae subgroup. However, the rate of adaptive evolution is not greater for male-biased genes than for female-biased or unbiased genes, which suggests that there are differences in sex-biased gene evolution between the two lineages.
Figures
Similar articles
-
Molecular evolution of sex-biased genes in Drosophila.Mol Biol Evol. 2004 Nov;21(11):2130-9. doi: 10.1093/molbev/msh223. Epub 2004 Jul 28. Mol Biol Evol. 2004. PMID: 15282334
-
Selection shapes turnover and magnitude of sex-biased expression in Drosophila gonads.BMC Evol Biol. 2019 Feb 20;19(1):60. doi: 10.1186/s12862-019-1377-4. BMC Evol Biol. 2019. PMID: 30786879 Free PMC article.
-
Evolution of hydra, a recently evolved testis-expressed gene with nine alternative first exons in Drosophila melanogaster.PLoS Genet. 2007 Jul;3(7):e107. doi: 10.1371/journal.pgen.0030107. PLoS Genet. 2007. PMID: 17616977 Free PMC article.
-
Population and behaviour genetics of Drosophila ananassae.Genetica. 1996 May;97(3):321-9. doi: 10.1007/BF00055318. Genetica. 1996. PMID: 9081860 Review.
-
Patterns of inversion polymorphism in three species of the Drosophila melanogaster species group.Indian J Exp Biol. 2001 Jul;39(7):611-22. Indian J Exp Biol. 2001. PMID: 12019752 Review.
Cited by
-
The coevolutionary period of Wolbachia pipientis infecting Drosophila ananassae and its impact on the evolution of the host germline stem cell regulating genes.Mol Biol Evol. 2014 Sep;31(9):2457-71. doi: 10.1093/molbev/msu204. Epub 2014 Jun 28. Mol Biol Evol. 2014. PMID: 24974378 Free PMC article.
-
Weighing the evidence for adaptation at the molecular level.Trends Genet. 2011 Sep;27(9):343-9. doi: 10.1016/j.tig.2011.06.003. Epub 2011 Jul 19. Trends Genet. 2011. PMID: 21775012 Free PMC article.
-
Genomic resources for multiple species in the Drosophila ananassae species group.Fly (Austin). 2013 Jan-Mar;7(1):47-57. doi: 10.4161/fly.22353. Fly (Austin). 2013. PMID: 23639891 Free PMC article.
-
Positive selection in nucleoporins challenges constraints on early expressed genes in Drosophila development.Genome Biol Evol. 2013;5(11):2231-41. doi: 10.1093/gbe/evt156. Genome Biol Evol. 2013. PMID: 24171912 Free PMC article.
-
Inter- and intraspecific variation in Drosophila genes with sex-biased expression.Int J Evol Biol. 2012;2012:963976. doi: 10.1155/2012/963976. Epub 2012 Jan 12. Int J Evol Biol. 2012. PMID: 22315698 Free PMC article.
References
-
- Zhang Z, Parsch J. Positive correlation between evolutionary rate and recombination rate in Drosophila genes with male-biased expression. Drosophila. 2005;22:1945–1947. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

