Maximizing RNA yield from archival renal tumors and optimizing gene expression analysis
- PMID: 20008123
- PMCID: PMC2902272
- DOI: 10.1177/1087057109355059
Maximizing RNA yield from archival renal tumors and optimizing gene expression analysis
Abstract
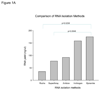
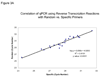
Formalin-fixed, paraffin-embedded tissues are widely available for gene expression analysis using TaqMan PCR. Five methods, including 4 commercial kits, for recovering RNA from paraffin-embedded renal tumor tissue were compared. The MasterPure kit from Epicentre produced the highest RNA yield. However, the difference in RNA yield between the kit from Epicenter and Invitrogen's TRIzol method was not significant. Using the top 3 RNA isolation methods, the manufacturers' protocols were modified to include an overnight Proteinase K digestion. Overnight protein digestion resulted in a significant increase in RNA yield. To optimize the reverse transcription reaction, conventional reverse transcription with random oligonucleotide primers was compared to reverse transcription using primers specific for genes of interest. Reverse transcription using gene-specific primers significantly increased the quantity of cDNA detectable by TaqMan PCR. Therefore, expression profiling of formalin-fixed, paraffin-embedded tissue using TaqMan qPCR can be optimized by using the MasterPure RNA isolation kit modified to include an overnight Proteinase K digestion and gene-specific primers during the reverse transcription.
Figures




Similar articles
-
Expression profiling of archival renal tumors by quantitative PCR to validate prognostic markers.Biotechniques. 2007 Nov;43(5):639-40, 642-3, 647. doi: 10.2144/000112562. Biotechniques. 2007. PMID: 18072593
-
Illumina whole-genome complementary DNA-mediated annealing, selection, extension and ligation platform: assessing its performance in formalin-fixed, paraffin-embedded samples and identifying invasion pattern-related genes in oral squamous cell carcinoma.Hum Pathol. 2011 Dec;42(12):1911-22. doi: 10.1016/j.humpath.2011.02.011. Epub 2011 Jun 17. Hum Pathol. 2011. PMID: 21683979
-
Optimization of the method of RNA isolation from paraffin blocks to assess gene expression in breast cancer.Pol J Pathol. 2008;59(2):85-91. Pol J Pathol. 2008. PMID: 18669173
-
Unlocking the archive--gene expression in paraffin-embedded tissue.J Pathol. 2001 Sep;195(1):66-71. doi: 10.1002/1096-9896(200109)195:1<66::AID-PATH921>3.0.CO;2-F. J Pathol. 2001. PMID: 11568892 Review.
-
Tissue handling and specimen preparation in surgical pathology: issues concerning the recovery of nucleic acids from formalin-fixed, paraffin-embedded tissue.Arch Pathol Lab Med. 2008 Dec;132(12):1929-35. doi: 10.5858/132.12.1929. Arch Pathol Lab Med. 2008. PMID: 19061293 Review.
Cited by
-
A comparison of Ku0063794, a dual mTORC1 and mTORC2 inhibitor, and temsirolimus in preclinical renal cell carcinoma models.PLoS One. 2013;8(1):e54918. doi: 10.1371/journal.pone.0054918. Epub 2013 Jan 22. PLoS One. 2013. PMID: 23349989 Free PMC article.
-
Evaluation of microRNA expression in patient bone marrow aspirate slides.PLoS One. 2012;7(8):e42951. doi: 10.1371/journal.pone.0042951. Epub 2012 Aug 13. PLoS One. 2012. PMID: 22912766 Free PMC article.
-
Genes involved in prostate cancer progression determine MRI visibility.Theranostics. 2018 Feb 12;8(7):1752-1765. doi: 10.7150/thno.23180. eCollection 2018. Theranostics. 2018. PMID: 29556354 Free PMC article.
-
Recommendations for mRNA analysis of micro-dissected glomerular tufts from paraffin-embedded human kidney biopsy samples.BMC Mol Biol. 2018 Mar 13;19(1):2. doi: 10.1186/s12867-018-0103-x. BMC Mol Biol. 2018. PMID: 29534701 Free PMC article.
-
Evaluation of a high-throughput, microfluidics platform for performing TaqMan™ qPCR using formalin-fixed paraffin-embedded tumors.Bioanalysis. 2013 Jul;5(13):1623-33. doi: 10.4155/bio.13.125. Bioanalysis. 2013. PMID: 23822126 Free PMC article.
References
-
- Gnanapragasam V. Unlocking the molecular archive: the emerging use of formalin-fixed paraffin-embedded tissue for biomarker research in urological cancer. BJU Int. 2009 [Epub ahead of print] - PubMed
-
- Glenn ST, Jones CA, Liang P, Kaushik D, Gross KW, Kim HL. Expression profiling of archival renal tumors by quantitative PCR to validate prognostic markers. Biotechniques. 2007;43:639–640. 642-633, 647. - PubMed
-
- Chung JY, Braunschweig T, Hewitt SM. Optimization of recovery of RNA from formalin-fixed, paraffin-embedded tissue. Diagn Mol Pathol. 2006;15:229–236. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

