Cooperative nature of gating transitions in K(+) channels as seen from dynamic importance sampling calculations
- PMID: 19950367
- PMCID: PMC2822122
- DOI: 10.1002/prot.22632
Cooperative nature of gating transitions in K(+) channels as seen from dynamic importance sampling calculations
Abstract
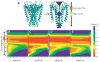
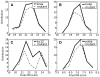
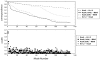
The growing dataset of K(+) channel x-ray structures provides an excellent opportunity to begin a detailed molecular understanding of voltage-dependent gating. These structures, while differing in sequence, represent either a stable open or closed state. However, an understanding of the molecular details of gating will require models for the transitions and experimentally testable predictions for the gating transition. To explore these ideas, we apply dynamic importance sampling to a set of homology models for the molecular conformations of K(+) channels for four different sets of sequences and eight different states. In our results, we highlight the importance of particular residues upstream from the Pro-Val-Pro (PVP) region to the gating transition. This supports growing evidence that the PVP region is important for influencing the flexibility of the S6 helix and thus the opening of the gating domain. The results further suggest how gating on the molecular level depends on intra-subunit motions to influence the cooperative behavior of all four subunits of the K(+) channel. We hypothesize that the gating process occurs in steps: first sidechain movement, then inter-S5-S6 subunit motions, and lastly the large-scale domain rearrangements.
Figures




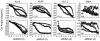
Similar articles
-
Dynamic interaction of S5 and S6 during voltage-controlled gating in a potassium channel.J Gen Physiol. 2001 Aug;118(2):157-70. doi: 10.1085/jgp.118.2.157. J Gen Physiol. 2001. PMID: 11479343 Free PMC article.
-
Cooperative transition between open and closed conformations in potassium channels.PLoS Comput Biol. 2008 Aug 29;4(8):e1000164. doi: 10.1371/journal.pcbi.1000164. PLoS Comput Biol. 2008. PMID: 18769593 Free PMC article.
-
Gating of voltage-dependent potassium channels.Prog Biophys Mol Biol. 2001;75(3):165-99. doi: 10.1016/s0079-6107(01)00006-2. Prog Biophys Mol Biol. 2001. PMID: 11376798 Review.
-
Proline-induced hinges in transmembrane helices: possible roles in ion channel gating.Proteins. 2001 Aug 1;44(2):63-72. doi: 10.1002/prot.1073. Proteins. 2001. PMID: 11391769
-
Structural changes during ion channel gating.Trends Neurosci. 2004 Jun;27(6):298-302. doi: 10.1016/j.tins.2004.04.004. Trends Neurosci. 2004. PMID: 15165732 Review.
Cited by
-
Being flexible: the voltage-controllable activation gate of kv channels.Front Pharmacol. 2012 Sep 13;3:168. doi: 10.3389/fphar.2012.00168. eCollection 2012. Front Pharmacol. 2012. PMID: 22993508 Free PMC article.
-
Golgi anti-apoptotic proteins are highly conserved ion channels that affect apoptosis and cell migration.J Biol Chem. 2015 May 1;290(18):11785-801. doi: 10.1074/jbc.M115.637306. Epub 2015 Feb 24. J Biol Chem. 2015. PMID: 25713081 Free PMC article.
-
Hydrophobic plug functions as a gate in voltage-gated proton channels.Proc Natl Acad Sci U S A. 2014 Jan 14;111(2):E273-82. doi: 10.1073/pnas.1318018111. Epub 2013 Dec 30. Proc Natl Acad Sci U S A. 2014. PMID: 24379371 Free PMC article.
-
Probing the energy landscape of activation gating of the bacterial potassium channel KcsA.PLoS Comput Biol. 2013;9(5):e1003058. doi: 10.1371/journal.pcbi.1003058. Epub 2013 May 2. PLoS Comput Biol. 2013. PMID: 23658510 Free PMC article.
-
Mutations in the S6 gate isolate a late step in the activation pathway and reduce 4-AP sensitivity in shaker K(v) channel.Biophys J. 2014 Jan 7;106(1):134-44. doi: 10.1016/j.bpj.2013.11.025. Biophys J. 2014. PMID: 24411245 Free PMC article.
References
-
- Hille B. Ion Channels of Excitable Membranes. 3. Sinauer; Sunderland, MA: 2001.
-
- Zhou Y, Morais-Cabral JH, Kaufman A, MacKinnon R. Chemistry of ion coordination and hydration revealed by a K+ channel-Fab complex at 2.0 Å resolution. Nature. 2001;414:43–48. - PubMed
-
- Jiang Y Lee A, Chen J, Cadene M, Chait BT, MacKinnon R. Crystal Structure and Mechanism of a Calcium-Gated Potassium Channel. Nature. 2002;417:515–522. - PubMed
-
- Jiang Y, Lee A, Chen J, Ruta V, Cadene M, Chait BT, MacKinnon R. X-ray structure of a voltage-dependent K+ channel. Nature. 2003;423:33–41. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

