Fusion-deficient insertion mutants of herpes simplex virus type 1 glycoprotein B adopt the trimeric postfusion conformation
- PMID: 19939928
- PMCID: PMC2812406
- DOI: 10.1128/JVI.01791-09
Fusion-deficient insertion mutants of herpes simplex virus type 1 glycoprotein B adopt the trimeric postfusion conformation
Abstract
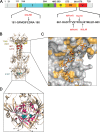
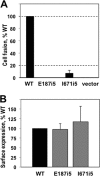
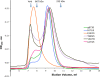
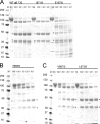
Glycoprotein B (gB) enables the fusion of viral and cell membranes during entry of herpesviruses. However, gB alone is insufficient for membrane fusion; the gH/gL heterodimer is also required. The crystal structure of the herpes simplex virus type 1 (HSV-1) gB ectodomain, gB730, has demonstrated similarities between gB and other viral fusion proteins, leading to the hypothesis that gB is a fusogen, presumably directly involved in bringing the membranes together by refolding from its initial or prefusion form to its final or postfusion form. The only available crystal structure likely represents the postfusion form of gB; the prefusion form has not yet been determined. Previously, a panel of HSV-1 gB mutants was generated by using random 5-amino-acid-linker insertion mutagenesis. Several mutants were unable to mediate cell-cell fusion despite being expressed on the cell surface. Mapping of the insertion sites onto the crystal structure of gB730 suggested that several insertions might not be accommodated in the postfusion form. Thus, we hypothesized that some insertion mutants were nonfunctional due to being "trapped" in a prefusion form. Here, we generated five insertion mutants as soluble ectodomains and characterized them biochemically. We show that the ectodomains of all five mutants assume conformations similar to that of the wild-type gB730. Four mutants have biochemical properties and overall structures that are indistinguishable from those of the wild-type gB730. We conclude that these mutants undergo only minor local conformational changes to relieve the steric strain resulting from the presence of 5 extra amino acids. Interestingly, one mutant, while able to adopt the overall postfusion structure, displays significant conformational differences in the vicinity of fusion loops, relative to wild-type gB730. Moreover, this mutant has a diminished ability to associate with liposomes, suggesting that the fusion loops in this mutant have decreased functional activity. We propose that these insertions cause a fusion-deficient phenotype not by preventing conversion of gB to a postfusion-like conformation but rather by interfering with other gB functions.
Figures




Similar articles
-
Structure-Based Mutations in the Herpes Simplex Virus 1 Glycoprotein B Ectodomain Arm Impart a Slow-Entry Phenotype.mBio. 2017 May 16;8(3):e00614-17. doi: 10.1128/mBio.00614-17. mBio. 2017. PMID: 28512095 Free PMC article.
-
Natural Selection of Glycoprotein B Mutations That Rescue the Small-Plaque Phenotype of a Fusion-Impaired Herpes Simplex Virus Mutant.mBio. 2018 Oct 16;9(5):e01948-18. doi: 10.1128/mBio.01948-18. mBio. 2018. PMID: 30327436 Free PMC article.
-
Extensive mutagenesis of the HSV-1 gB ectodomain reveals remarkable stability of its postfusion form.J Mol Biol. 2013 Jun 12;425(11):2056-2071. doi: 10.1016/j.jmb.2013.03.001. Epub 2013 Mar 13. J Mol Biol. 2013. PMID: 23500487 Free PMC article.
-
Herpesvirus membrane fusion - a team effort.Curr Opin Struct Biol. 2020 Jun;62:112-120. doi: 10.1016/j.sbi.2019.12.004. Epub 2020 Jan 11. Curr Opin Struct Biol. 2020. PMID: 31935542 Review.
-
Structures of vesicular stomatitis virus glycoprotein: membrane fusion revisited.Cell Mol Life Sci. 2008 Jun;65(11):1716-28. doi: 10.1007/s00018-008-7534-3. Cell Mol Life Sci. 2008. PMID: 18345480 Free PMC article. Review.
Cited by
-
The membrane-proximal region (MPR) of herpes simplex virus gB regulates association of the fusion loops with lipid membranes.mBio. 2012 Nov 20;3(6):e00429-12. doi: 10.1128/mBio.00429-12. mBio. 2012. PMID: 23170000 Free PMC article.
-
Structural basis of local, pH-dependent conformational changes in glycoprotein B from herpes simplex virus type 1.J Virol. 2010 Dec;84(24):12924-33. doi: 10.1128/JVI.01750-10. Epub 2010 Oct 13. J Virol. 2010. PMID: 20943984 Free PMC article.
-
Selective Degradation of Host RNA Polymerase II Transcripts by Influenza A Virus PA-X Host Shutoff Protein.PLoS Pathog. 2016 Feb 5;12(2):e1005427. doi: 10.1371/journal.ppat.1005427. eCollection 2016 Feb. PLoS Pathog. 2016. PMID: 26849127 Free PMC article.
-
Residues within the C-terminal arm of the herpes simplex virus 1 glycoprotein B ectodomain contribute to its refolding during the fusion step of virus entry.J Virol. 2012 Jun;86(12):6386-93. doi: 10.1128/JVI.00104-12. Epub 2012 Apr 4. J Virol. 2012. PMID: 22491468 Free PMC article.
-
Membrane requirement for folding of the herpes simplex virus 1 gB cytodomain suggests a unique mechanism of fusion regulation.J Virol. 2012 Aug;86(15):8171-84. doi: 10.1128/JVI.00932-12. Epub 2012 May 23. J Virol. 2012. PMID: 22623783 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

