Fission yeast Iec1-ino80-mediated nucleosome eviction regulates nucleotide and phosphate metabolism
- PMID: 19933844
- PMCID: PMC2812234
- DOI: 10.1128/MCB.01117-09
Fission yeast Iec1-ino80-mediated nucleosome eviction regulates nucleotide and phosphate metabolism
Abstract
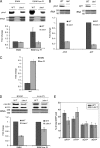
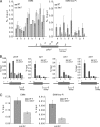
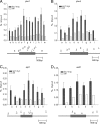
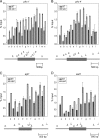
Ino80 is an ATP-dependent nucleosome-remodeling enzyme involved in transcription, replication, and the DNA damage response. Here, we characterize the fission yeast Ino80 and find that it is essential for cell viability. We show that the Ino80 complex from fission yeast mediates ATP-dependent nucleosome remodeling in vitro. The purification of the Ino80-associated complex identified a highly conserved complex and the presence of a novel zinc finger protein with similarities to the mammalian transcriptional regulator Yin Yang 1 (YY1) and other members of the GLI-Krüppel family of proteins. Deletion of this Iec1 protein or the Ino80 complex subunit arp8, ies6, or ies2 causes defects in DNA damage repair, the response to replication stress, and nucleotide metabolism. We show that Iec1 is important for the correct expression of genes involved in nucleotide metabolism, including the ribonucleotide reductase subunit cdc22 and phosphate- and adenine-responsive genes. We find that Ino80 is recruited to a large number of promoter regions on phosphate starvation, including those of phosphate- and adenine-responsive genes that depend on Iec1 for correct expression. Iec1 is required for the binding of Ino80 to target genes and subsequent histone loss at the promoter and throughout the body of these genes on phosphate starvation. This suggests that the Iec1-Ino80 complex promotes transcription through nucleosome eviction.
Figures




Similar articles
-
An essential role for the Ino80 chromatin remodeling complex in regulation of gene expression during cellular quiescence.Chromosome Res. 2023 Apr 12;31(2):14. doi: 10.1007/s10577-023-09723-x. Chromosome Res. 2023. PMID: 37043046 Free PMC article.
-
The INO80 complex activates the transcription of S-phase genes in a cell cycle-regulated manner.FEBS J. 2018 Oct;285(20):3870-3881. doi: 10.1111/febs.14640. Epub 2018 Sep 6. FEBS J. 2018. PMID: 30134042
-
INO80 represses osmostress induced gene expression by resetting promoter proximal nucleosomes.Nucleic Acids Res. 2017 Apr 20;45(7):3752-3766. doi: 10.1093/nar/gkw1292. Nucleic Acids Res. 2017. PMID: 28025392 Free PMC article.
-
The INO80 remodeller in transcription, replication and repair.Philos Trans R Soc Lond B Biol Sci. 2017 Oct 5;372(1731):20160290. doi: 10.1098/rstb.2016.0290. Philos Trans R Soc Lond B Biol Sci. 2017. PMID: 28847827 Free PMC article. Review.
-
A conserved role of the RSC chromatin remodeler in the establishment of nucleosome-depleted regions.Curr Genet. 2017 May;63(2):187-193. doi: 10.1007/s00294-016-0642-y. Epub 2016 Aug 24. Curr Genet. 2017. PMID: 27558480 Free PMC article. Review.
Cited by
-
Beyond the histone tail: acetylation at the nucleosome dyad commands transcription.Nucleus. 2013 Sep-Oct;4(5):343-8. doi: 10.4161/nucl.26051. Epub 2013 Aug 9. Nucleus. 2013. PMID: 23941995 Free PMC article.
-
Fitness profiling links topoisomerase II regulation of centromeric integrity to doxorubicin resistance in fission yeast.Sci Rep. 2015 Feb 11;5:8400. doi: 10.1038/srep08400. Sci Rep. 2015. PMID: 25669599 Free PMC article.
-
Hap2-Ino80-facilitated transcription promotes de novo establishment of CENP-A chromatin.Genes Dev. 2020 Feb 1;34(3-4):226-238. doi: 10.1101/gad.332536.119. Epub 2020 Jan 9. Genes Dev. 2020. PMID: 31919190 Free PMC article.
-
An essential role for the Ino80 chromatin remodeling complex in regulation of gene expression during cellular quiescence.Chromosome Res. 2023 Apr 12;31(2):14. doi: 10.1007/s10577-023-09723-x. Chromosome Res. 2023. PMID: 37043046 Free PMC article.
-
Epigenetic Regulation of Chromatin States in Schizosaccharomyces pombe.Cold Spring Harb Perspect Biol. 2015 Jul 1;7(7):a018770. doi: 10.1101/cshperspect.a018770. Cold Spring Harb Perspect Biol. 2015. PMID: 26134317 Free PMC article. Review.
References
-
- Altschul, S. F., and E. V. Koonin. 1998. Iterated profile searches with PSI-BLAST—a tool for discovery in protein databases. Trends Biochem. Sci. 23:444-447. - PubMed
-
- Bähler, J., J. Q. Wu, M. S. Longtine, N. G. Shah, A. McKenzie, A. B. Steever, A. Wach, P. Philippsen, and J. R. Pringle. 1998. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast 14:943-951. - PubMed
-
- Barbaric, S., T. Luckenbach, A. Schmid, D. Blaschke, W. Horz, and P. Korber. 2007. Redundancy of chromatin remodeling pathways for the induction of the yeast PHO5 promoter in vivo. J. Biol. Chem. 282:27610-27621. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- BB/F020236/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BBS/E/B/0000H125/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- G0701175/MRC_/Medical Research Council/United Kingdom
- (BB/F020236/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
