Functional analysis of conserved aromatic amino acids in the discoidin domain of Paenibacillus beta-1,3-glucanase
- PMID: 19930717
- PMCID: PMC2789033
- DOI: 10.1186/1475-2859-8-62
Functional analysis of conserved aromatic amino acids in the discoidin domain of Paenibacillus beta-1,3-glucanase
Abstract
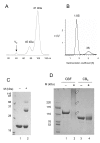
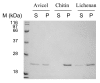
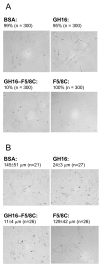
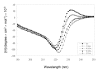
The 190-kDa Paenibacillus beta-1,3-glucanase (LamA) contains a catalytic module of the glycoside hydrolase family 16 (GH16) and several auxiliary domains. Of these, a discoidin domain (DS domain), present in both eukaryotic and prokaryotic proteins with a wide variety of functions, exists at the carboxyl-terminus. To better understand the bacterial DS domain in terms of its structure and function, this domain alone was expressed in Escherichia coli and characterized. The results indicate that the DS domain binds various polysaccharides and enhances the biological activity of the GH16 module on composite substrates. We also investigated the importance of several conserved aromatic residues in the domain's stability and substrate-binding affinity. Both were affected by mutations of these residues; however, the effect on protein stability was more notable. In particular, the forces contributed by a sandwiched triad (W1688, R1756, and W1729) were critical for the presumable beta-sandwich fold.
Figures

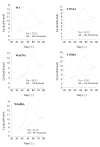

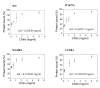
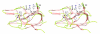
Similar articles
-
Recombinant production and characterization of full-length and truncated β-1,3-glucanase PglA from Paenibacillus sp. S09.BMC Biotechnol. 2013 Nov 28;13:105. doi: 10.1186/1472-6750-13-105. BMC Biotechnol. 2013. PMID: 24283345 Free PMC article.
-
Biochemical and genetic properties of Paenibacillus glycosyl hydrolase having chitosanase activity and discoidin domain.J Biol Chem. 2002 Apr 26;277(17):14695-702. doi: 10.1074/jbc.M108660200. Epub 2002 Feb 19. J Biol Chem. 2002. PMID: 11854270
-
The discoidin domain of Bacillus circulans β-galactosidase plays an essential role in repressing galactooligosaccharide production.Biosci Biotechnol Biochem. 2013;77(1):73-9. doi: 10.1271/bbb.120583. Epub 2013 Jan 7. Biosci Biotechnol Biochem. 2013. PMID: 23291776
-
Structural insights into substrate recognition and catalysis by glycoside hydrolase family 87 α-1,3-glucanase from Paenibacillus glycanilyticus FH11.FEBS J. 2020 Jun;287(12):2524-2543. doi: 10.1111/febs.15161. Epub 2019 Dec 19. FEBS J. 2020. PMID: 31788942
-
Structural similarities and functional diversity of eukaryotic discoidin-like domains.Biochim Biophys Acta. 2007 Sep;1774(9):1069-78. doi: 10.1016/j.bbapap.2007.07.007. Epub 2007 Jul 24. Biochim Biophys Acta. 2007. PMID: 17702679 Review.
Cited by
-
Functional Analysis of a Novel β-(1,3)-Glucanase from Corallococcus sp. Strain EGB Containing a Fascin-Like Module.Appl Environ Microbiol. 2017 Aug 1;83(16):e01016-17. doi: 10.1128/AEM.01016-17. Print 2017 Aug 15. Appl Environ Microbiol. 2017. PMID: 28625980 Free PMC article.
-
α-1,3-Glucanase: present situation and prospect of research.World J Microbiol Biotechnol. 2016 Feb;32(2):30. doi: 10.1007/s11274-015-1977-0. Epub 2016 Jan 9. World J Microbiol Biotechnol. 2016. PMID: 26748807 Review.
-
Recombinant production and characterization of full-length and truncated β-1,3-glucanase PglA from Paenibacillus sp. S09.BMC Biotechnol. 2013 Nov 28;13:105. doi: 10.1186/1472-6750-13-105. BMC Biotechnol. 2013. PMID: 24283345 Free PMC article.
-
Putative chitin synthases from Branchiostoma floridae show extracellular matrix-related domains and mosaic structures.Genomics Proteomics Bioinformatics. 2012 Aug;10(4):197-207. doi: 10.1016/j.gpb.2012.07.003. Epub 2012 Jul 31. Genomics Proteomics Bioinformatics. 2012. PMID: 23084775 Free PMC article.
References
-
- Hidai C, Zupancic T, Penta K, Mikhail A, Kawana M, Quertermous EE, Aoka Y, Fukagawa M, Matsui Y, Platika D, Auerbach R, Hogan BLM, Snodgrass R, Quertermous T. Cloning and characterization of developmental endothelial locus-1: an embryonic endothelial cell protein that binds the αvβ 3 integrin receptor. Genes Dev. 1998;12:21–33. doi: 10.1101/gad.12.1.21. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

