Nap1 and Chz1 have separate Htz1 nuclear import and assembly functions
- PMID: 19929865
- PMCID: PMC2907061
- DOI: 10.1111/j.1600-0854.2009.01010.x
Nap1 and Chz1 have separate Htz1 nuclear import and assembly functions
Abstract
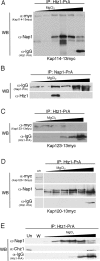
We analyzed the nuclear import and regulation of the yeast histone variant Htz1 (H2A.Z), and the role of histone chaperones Nap1 and Chz1 in this process. Copurification suggested that Htz1 and H2B dimerized in the cytoplasm prior to import. Like H2B, Htz1 contained a nuclear localization signal (NLS) in its N-terminus that is recognized by multiple karyopherins (also called importins), indicating multiple transport pathways into the nucleus. However, Kap114 and Kap123 appeared to play the major role in Htz1 import. We also identified a role for Nap1 in the import of Htz1/H2B heterodimers, and Nap1 formed a RanGTP-insensitive import complex with Htz1/H2B and Kap114. Nap1 was necessary for maintaining a soluble pool of Htz1, indicating that its chaperone function may be important for the dynamic exchange of histones within nucleosomes. In contrast, Chz1 was imported by a distinct import pathway, and Chz1 did not appear to interact with Htz1 in the cytoplasm. Genetic analysis indicated that NAP1 has a function in the absence of HTZ1 that is not shared with CHZ1. This provides further evidence that the histone chaperones Nap1 and Chz1 have separate Htz1-dependent and -independent functions.
Figures



Similar articles
-
Distinct roles for histone chaperones in the deposition of Htz1 in chromatin.Biosci Rep. 2014 Sep 19;34(5):e00139. doi: 10.1042/BSR20140092. Biosci Rep. 2014. PMID: 25338502 Free PMC article.
-
Histone chaperone Chz1 facilitates the disfavouring property of Spt16 to H2A.Z-containing genes in Saccharomyces cerevisiae.Biochem J. 2014 Jun 15;460(3):387-97. doi: 10.1042/BJ20140186. Biochem J. 2014. PMID: 24707933
-
Nuclear import of histone H2A and H2B is mediated by a network of karyopherins.J Cell Biol. 2001 Apr 16;153(2):251-62. doi: 10.1083/jcb.153.2.251. J Cell Biol. 2001. PMID: 11309407 Free PMC article.
-
Nuclear import of histones.Biochem Soc Trans. 2020 Dec 18;48(6):2753-2767. doi: 10.1042/BST20200572. Biochem Soc Trans. 2020. PMID: 33300986 Free PMC article. Review.
-
Histone chaperones link histone nuclear import and chromatin assembly.Biochim Biophys Acta. 2013 Mar-Apr;1819(3-4):277-89. Biochim Biophys Acta. 2013. PMID: 24459730 Review.
Cited by
-
Molecular basis of RanGTP-activated nucleosome assembly with Histones H2A-H2B bound to Importin-9.bioRxiv [Preprint]. 2023 Jan 28:2023.01.27.525896. doi: 10.1101/2023.01.27.525896. bioRxiv. 2023. Update in: Structure. 2023 Aug 3;31(8):903-911.e3. doi: 10.1016/j.str.2023.06.001 PMID: 36747879 Free PMC article. Updated. Preprint.
-
Histone chaperone networks shaping chromatin function.Nat Rev Mol Cell Biol. 2017 Mar;18(3):141-158. doi: 10.1038/nrm.2016.159. Epub 2017 Jan 5. Nat Rev Mol Cell Biol. 2017. PMID: 28053344 Free PMC article. Review.
-
Interaction with the histone chaperone Vps75 promotes nuclear localization and HAT activity of Rtt109 in vivo.Traffic. 2011 Jul;12(7):826-39. doi: 10.1111/j.1600-0854.2011.01202.x. Epub 2011 May 5. Traffic. 2011. PMID: 21463458 Free PMC article.
-
Individual lysine acetylations on the N terminus of Saccharomyces cerevisiae H2A.Z are highly but not differentially regulated.J Biol Chem. 2010 Dec 17;285(51):39855-65. doi: 10.1074/jbc.M110.185967. Epub 2010 Oct 14. J Biol Chem. 2010. PMID: 20952395 Free PMC article.
-
Chaperoning RPA during DNA metabolism.Curr Genet. 2019 Aug;65(4):857-864. doi: 10.1007/s00294-019-00945-3. Epub 2019 Feb 22. Curr Genet. 2019. PMID: 30796471 Review.
References
-
- Luger K. Structure and dynamic behavior of nucleosomes. Current opinion in genetics & development. 2003;13(2):127–135. - PubMed
-
- Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389(6648):251–260. - PubMed
-
- Guillemette B, Gaudreau L. Reuniting the contrasting functions of H2A.Z. Biochemistry and cell biology = Biochimie et biologie cellulaire. 2006;84(4):528–535. - PubMed
-
- Raisner RM, Madhani HD. Patterning chromatin: form and function for H2A.Z variant nucleosomes. Current opinion in genetics & development. 2006;16(2):119–124. - PubMed
-
- Zlatanova J, Thakar A. H2A.Z: view from the top. Structure. 2008;16(2):166–179. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

