Calling International Rescue: knowledge lost in literature and data landslide!
- PMID: 19929850
- PMCID: PMC2805925
- DOI: 10.1042/BJ20091474
Calling International Rescue: knowledge lost in literature and data landslide!
Abstract
We live in interesting times. Portents of impending catastrophe pervade the literature, calling us to action in the face of unmanageable volumes of scientific data. But it isn't so much data generation per se, but the systematic burial of the knowledge embodied in those data that poses the problem: there is so much information available that we simply no longer know what we know, and finding what we want is hard - too hard. The knowledge we seek is often fragmentary and disconnected, spread thinly across thousands of databases and millions of articles in thousands of journals. The intellectual energy required to search this array of data-archives, and the time and money this wastes, has led several researchers to challenge the methods by which we traditionally commit newly acquired facts and knowledge to the scientific record. We present some of these initiatives here - a whirlwind tour of recent projects to transform scholarly publishing paradigms, culminating in Utopia and the Semantic Biochemical Journal experiment. With their promises to provide new ways of interacting with the literature, and new and more powerful tools to access and extract the knowledge sequestered within it, we ask what advances they make and what obstacles to progress still exist? We explore these questions, and, as you read on, we invite you to engage in an experiment with us, a real-time test of a new technology to rescue data from the dormant pages of published documents. We ask you, please, to read the instructions carefully. The time has come: you may turn over your papers...
Figures
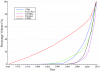







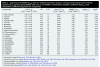




Similar articles
-
[Publications for rheumatology in new forms. What will come--what will remain?].Z Rheumatol. 2006 Feb;65(1):5. doi: 10.1007/s00393-006-0043-6. Z Rheumatol. 2006. PMID: 16804992 German. No abstract available.
-
The attention economy and the EMBS.IEEE Eng Med Biol Mag. 2004 May-Jun;23(3):5-6. IEEE Eng Med Biol Mag. 2004. PMID: 15354987 No abstract available.
-
Folic acid supplementation and malaria susceptibility and severity among people taking antifolate antimalarial drugs in endemic areas.Cochrane Database Syst Rev. 2022 Feb 1;2(2022):CD014217. doi: 10.1002/14651858.CD014217. Cochrane Database Syst Rev. 2022. PMID: 36321557 Free PMC article.
-
e-Science and its implications.Philos Trans A Math Phys Eng Sci. 2003 Aug 15;361(1809):1809-25. doi: 10.1098/rsta.2003.1224. Philos Trans A Math Phys Eng Sci. 2003. PMID: 12952686 Review.
-
Research by retrieving experiments.Cell Cycle. 2007 Jun 1;6(11):1277-83. doi: 10.4161/cc.6.11.4350. Epub 2007 Jun 26. Cell Cycle. 2007. PMID: 17525526 Review.
Cited by
-
Data Incompleteness May form a Hard-to-Overcome Barrier to Decoding Life's Mechanism.Biology (Basel). 2022 Aug 12;11(8):1208. doi: 10.3390/biology11081208. Biology (Basel). 2022. PMID: 36009835 Free PMC article.
-
Toward an interactive article: integrating journals and biological databases.BMC Bioinformatics. 2011 May 19;12:175. doi: 10.1186/1471-2105-12-175. BMC Bioinformatics. 2011. PMID: 21595960 Free PMC article.
-
Teaching computers to read the pharmacogenomics literature ... so you don't have to.Pharmacogenomics. 2010 Apr;11(4):515-8. doi: 10.2217/pgs.10.48. Pharmacogenomics. 2010. PMID: 20350132 Free PMC article. No abstract available.
-
Openness and trust in data-intensive science: the case of biocuration.Med Health Care Philos. 2020 Sep;23(3):497-504. doi: 10.1007/s11019-020-09960-5. Med Health Care Philos. 2020. PMID: 32524312 Free PMC article.
-
Drug design for ever, from hype to hope.J Comput Aided Mol Des. 2012 Jan;26(1):137-50. doi: 10.1007/s10822-011-9519-9. Epub 2012 Jan 18. J Comput Aided Mol Des. 2012. PMID: 22252446 Free PMC article.
References
-
- Roos D. Bioinformatics: trying to swim in a sea of data. Science. 2001;291:1260–1261. - PubMed
-
- Gerhold D., Rushmore T., Caskey C. T. DNA chips: promising toys have become powerful tools. Trends Biol. Sci. 1999;24:168–173. - PubMed
-
- Andrade M., Sander C. Bioinformatics: from genome data to biological knowledge. Curr. Opin. Biotechnol. 1997;8:675–683. - PubMed
-
- Hess K. R., Zhang W., Baggerly K. A., Stivers D. N., Coombes K. R., Zhang W. Micro-arrays: handling the deluge of data and extracting reliable information. Trends Biotechnol. 2001;19:463–468. - PubMed
-
- Editorial. Prepare for the deluge. Nat. Biotechnol. 2008;26:1099. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

