Coordinating DNA replication by means of priming loop and differential synthesis rate
- PMID: 19924126
- PMCID: PMC2896039
- DOI: 10.1038/nature08611
Coordinating DNA replication by means of priming loop and differential synthesis rate
Abstract
Genomic DNA is replicated by two DNA polymerase molecules, one of which works in close association with the helicase to copy the leading-strand template in a continuous manner while the second copies the already unwound lagging-strand template in a discontinuous manner through the synthesis of Okazaki fragments. Considering that the lagging-strand polymerase has to recycle after the completion of every Okazaki fragment through the slow steps of primer synthesis and hand-off to the polymerase, it is not understood how the two strands are synthesized with the same net rate. Here we show, using the T7 replication proteins, that RNA primers are made 'on the fly' during ongoing DNA synthesis and that the leading-strand T7 replisome does not pause during primer synthesis, contrary to previous reports. Instead, the leading-strand polymerase remains limited by the speed of the helicase; it therefore synthesizes DNA more slowly than the lagging-strand polymerase. We show that the primase-helicase T7 gp4 maintains contact with the priming sequence during ongoing DNA synthesis; the nascent lagging-strand template therefore organizes into a priming loop that keeps the primer in physical proximity to the replication complex. Our findings provide three synergistic mechanisms of coordination: first, primers are made concomitantly with DNA synthesis; second, the priming loop ensures efficient primer use and hand-off to the polymerase; and third, the lagging-strand polymerase copies DNA faster, which allows it to keep up with leading-strand DNA synthesis overall.
Figures


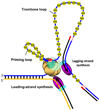
Comment in
-
DNA replication: prime-time looping.Nature. 2009 Dec 17;462(7275):854-5. doi: 10.1038/462854a. Nature. 2009. PMID: 20016583 No abstract available.
Similar articles
-
Discrete interactions between bacteriophage T7 primase-helicase and DNA polymerase drive the formation of a priming complex containing two copies of DNA polymerase.Biochemistry. 2013 Jun 11;52(23):4026-36. doi: 10.1021/bi400284j. Epub 2013 May 31. Biochemistry. 2013. PMID: 23675753 Free PMC article.
-
Binding Affinities among DNA Helicase-Primase, DNA Polymerase, and Replication Intermediates in the Replisome of Bacteriophage T7.J Biol Chem. 2016 Jan 15;291(3):1472-80. doi: 10.1074/jbc.M115.698233. Epub 2015 Nov 30. J Biol Chem. 2016. PMID: 26620561 Free PMC article.
-
Primer release is the rate-limiting event in lagging-strand synthesis mediated by the T7 replisome.Proc Natl Acad Sci U S A. 2016 May 24;113(21):5916-21. doi: 10.1073/pnas.1604894113. Epub 2016 May 9. Proc Natl Acad Sci U S A. 2016. PMID: 27162371 Free PMC article.
-
Choreography of bacteriophage T7 DNA replication.Curr Opin Chem Biol. 2011 Oct;15(5):580-6. doi: 10.1016/j.cbpa.2011.07.024. Epub 2011 Sep 9. Curr Opin Chem Biol. 2011. PMID: 21907611 Free PMC article. Review.
-
The Replication System of Bacteriophage T7.Enzymes. 2016;39:89-136. doi: 10.1016/bs.enz.2016.02.001. Epub 2016 Mar 28. Enzymes. 2016. PMID: 27241928 Review.
Cited by
-
Two mechanisms coordinate replication termination by the Escherichia coli Tus-Ter complex.Nucleic Acids Res. 2015 Jul 13;43(12):5924-35. doi: 10.1093/nar/gkv527. Epub 2015 May 24. Nucleic Acids Res. 2015. PMID: 26007657 Free PMC article.
-
Discrete interactions between bacteriophage T7 primase-helicase and DNA polymerase drive the formation of a priming complex containing two copies of DNA polymerase.Biochemistry. 2013 Jun 11;52(23):4026-36. doi: 10.1021/bi400284j. Epub 2013 May 31. Biochemistry. 2013. PMID: 23675753 Free PMC article.
-
Dynamic coupling between the motors of DNA replication: hexameric helicase, DNA polymerase, and primase.Curr Opin Chem Biol. 2011 Oct;15(5):595-605. doi: 10.1016/j.cbpa.2011.08.003. Epub 2011 Aug 22. Curr Opin Chem Biol. 2011. PMID: 21865075 Free PMC article. Review.
-
Excessive excision of correct nucleotides during DNA synthesis explained by replication hurdles.EMBO J. 2020 Mar 16;39(6):e103367. doi: 10.15252/embj.2019103367. Epub 2020 Feb 9. EMBO J. 2020. PMID: 32037587 Free PMC article.
-
The mechanism of gap creation by a multifunctional nuclease during base excision repair.Sci Adv. 2021 Jul 14;7(29):eabg0076. doi: 10.1126/sciadv.abg0076. Print 2021 Jul. Sci Adv. 2021. PMID: 34261654 Free PMC article.
References
-
- Benkovic SJ, Valentine AM, Salinas F. Replisome-mediated DNA replication. Annu Rev Biochem. 2001;70:181–208. - PubMed
-
- O'Donnell M. Replisome architecture and dynamics in Escherichia coli. J Biol Chem. 2006;281:10653–10656. - PubMed
-
- Stukenberg PT, Turner J, O'Donnell M. An explanation for lagging strand replication: polymerase hopping among DNA sliding clamps. Cell. 1994;78:877–887. - PubMed
-
- Frick DN, Richardson CC. DNA primases. Annu Rev Biochem. 2001;70:39–80. - PubMed
-
- Patel SS, Hingorani MM, Ng WM. The K318A mutant of bacteriophage T7 DNA primase-helicase protein is deficient in helicase but not primase activity and inhibits primase-helicase protein wild-type activities by heterooligomer formation. Biochemistry. 1994;33:7857–7868. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

