Refolding dynamics of stretched biopolymers upon force quench
- PMID: 19915145
- PMCID: PMC2787142
- DOI: 10.1073/pnas.0905764106
Refolding dynamics of stretched biopolymers upon force quench
Abstract
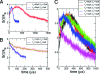
Single-molecule force spectroscopy methods can be used to generate folding trajectories of biopolymers from arbitrary regions of the folding landscape. We illustrate the complexity of the folding kinetics and generic aspects of the collapse of RNA and proteins upon force quench by using simulations of an RNA hairpin and theory based on the de Gennes model for homopolymer collapse. The folding time, tau(F), depends asymmetrically on deltaf(S) = f (S) - f (m) and deltaf (Q) = f (m) - f (Q) where f (S) (f (Q)) is the stretch (quench) force and f (m) is the transition midforce of the RNA hairpin. In accord with experiments, the relaxation kinetics of the molecular extension, R(t), occurs in three stages: A rapid initial decrease in the extension is followed by a plateau and finally, an abrupt reduction in R(t) occurs as the native state is approached. The duration of the plateau increases as lambda = tau (Q)/tau (F) decreases (where tau (Q) is the time in which the force is reduced from f (S) to f (Q)). Variations in the mechanisms of force-quench relaxation as lambda is altered are reflected in the experimentally measurable time-dependent entropy, which is computed directly from the folding trajectories. An analytical solution of the de Gennes model under tension reproduces the multistage stage kinetics in R(t). The prediction that the initial stages of collapse should also be a generic feature of polymers is validated by simulation of the kinetics of toroid (globule) formation in semiflexible (flexible) homopolymers in poor solvents upon quenching the force from a fully stretched state. Our findings give a unified explanation for multiple disparate experimental observations of protein folding.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


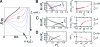

Similar articles
-
Multiple stepwise refolding of immunoglobulin domain I27 upon force quench depends on initial conditions.Proc Natl Acad Sci U S A. 2006 Jan 3;103(1):93-8. doi: 10.1073/pnas.0503758103. Epub 2005 Dec 22. Proc Natl Acad Sci U S A. 2006. PMID: 16373511 Free PMC article.
-
Mapping the energy landscape of biomolecules using single molecule force correlation spectroscopy: theory and applications.Biophys J. 2006 Jun 1;90(11):3827-41. doi: 10.1529/biophysj.105.075937. Epub 2006 Mar 13. Biophys J. 2006. PMID: 16533852 Free PMC article.
-
Forced-unfolding and force-quench refolding of RNA hairpins.Biophys J. 2006 May 15;90(10):3410-27. doi: 10.1529/biophysj.105.078030. Epub 2006 Feb 10. Biophys J. 2006. PMID: 16473903 Free PMC article.
-
Refolding upon force quench and pathways of mechanical and thermal unfolding of ubiquitin.Biophys J. 2007 Jan 15;92(2):547-61. doi: 10.1529/biophysj.106.087684. Epub 2006 Oct 27. Biophys J. 2007. PMID: 17071662 Free PMC article.
-
The development of single molecule force spectroscopy: from polymer biophysics to molecular machines.Q Rev Biophys. 2022 Aug 2;55:e9. doi: 10.1017/S0033583522000087. Q Rev Biophys. 2022. PMID: 35916314 Review.
Cited by
-
Beyond microscopic reversibility: Are observable non-equilibrium processes precisely reversible?J Chem Theory Comput. 2011 Aug 9;7(8):2520-2527. doi: 10.1021/ct200086k. J Chem Theory Comput. 2011. PMID: 21869866 Free PMC article.
-
Capturing transition paths and transition states for conformational rearrangements in the ribosome.Biophys J. 2014 Dec 16;107(12):2881-2890. doi: 10.1016/j.bpj.2014.10.022. Biophys J. 2014. PMID: 25517153 Free PMC article.
-
Biomolecular dynamics: order-disorder transitions and energy landscapes.Rep Prog Phys. 2012 Jul;75(7):076601. doi: 10.1088/0034-4885/75/7/076601. Epub 2012 Jun 28. Rep Prog Phys. 2012. PMID: 22790780 Free PMC article.
-
Effects of Preferential Counterion Interactions on the Specificity of RNA Folding.J Phys Chem Lett. 2018 Oct 4;9(19):5726-5732. doi: 10.1021/acs.jpclett.8b02086. Epub 2018 Sep 18. J Phys Chem Lett. 2018. PMID: 30211556 Free PMC article.
-
Reconstructing folding energy landscapes by single-molecule force spectroscopy.Annu Rev Biophys. 2014;43:19-39. doi: 10.1146/annurev-biophys-051013-022754. Annu Rev Biophys. 2014. PMID: 24895850 Free PMC article. Review.
References
-
- Thirumalai D, Hyeon C. RNA and protein folding: Common themes and variations. Biochemistry. 2005;44:4957–4970. - PubMed
-
- Onuchic JN, Wolynes PG. Theory of protein folding. Curr Opin Struct Biol. 2004;14:70–75. - PubMed
-
- Thirumalai D. From minimal models to real proteins: Time scales for protein folding kinetics. J Physique I. 1995;5:1457–1467.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

