Structural basis of the cross-reactivity of genetically related human anti-HIV-1 mAbs: implications for design of V3-based immunogens
- PMID: 19913488
- PMCID: PMC3683248
- DOI: 10.1016/j.str.2009.09.012
Structural basis of the cross-reactivity of genetically related human anti-HIV-1 mAbs: implications for design of V3-based immunogens
Abstract
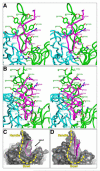
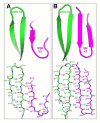
Human monoclonal antibodies 447-52D and 537-10D, both coded by the VH3 gene and specific for the third variable region (V3) of the HIV-1 gp120, were found to share antigen-binding structural elements including an elongated CDR H3 forming main-chain interactions with the N terminus of the V3 crown. However, water-mediated hydrogen bonds and a unique cation-pi sandwich stacking allow 447-52D to be broadly reactive with V3 containing both the GPGR and GPGQ crown motifs, while the deeper binding pocket and a buried Glu in the binding site of 537-10D limit its reactivity to only V3 containing the GPGR motif. Our results suggest that the design of immunogens for anti-V3 antibodies should avoid the Arg at the V3 crown, as GPGR-containing epitopes appear to select for B cells making antibodies of narrower specificity than V3 that carry Gln at this position.
Figures

Similar articles
-
The cross-clade neutralizing activity of a human monoclonal antibody is determined by the GPGR V3 motif of HIV type 1.AIDS Res Hum Retroviruses. 2004 Nov;20(11):1254-8. doi: 10.1089/aid.2004.20.1254. AIDS Res Hum Retroviruses. 2004. PMID: 15588347
-
Design of immunogens that present the crown of the HIV-1 V3 loop in a conformation competent to generate 447-52D-like antibodies.Biochem J. 2006 Nov 1;399(3):483-91. doi: 10.1042/BJ20060588. Biochem J. 2006. PMID: 16827663 Free PMC article.
-
Thermodynamic signatures of the antigen binding site of mAb 447-52D targeting the third variable region of HIV-1 gp120.Biochemistry. 2013 Sep 10;52(36):6249-57. doi: 10.1021/bi400645e. Epub 2013 Aug 23. Biochemistry. 2013. PMID: 23944979 Free PMC article.
-
Structural studies of human HIV-1 V3 antibodies.Hum Antibodies. 2005;14(3-4):73-80. Hum Antibodies. 2005. PMID: 16720977 Review.
-
NMR studies of V3 peptide complexes with antibodies suggest a mechanism for HIV-1 co-receptor selectivity.Curr Opin Drug Discov Devel. 2005 Sep;8(5):601-12. Curr Opin Drug Discov Devel. 2005. PMID: 16159022 Review.
Cited by
-
Human anti-V3 HIV-1 monoclonal antibodies encoded by the VH5-51/VL lambda genes define a conserved antigenic structure.PLoS One. 2011;6(12):e27780. doi: 10.1371/journal.pone.0027780. Epub 2011 Dec 2. PLoS One. 2011. PMID: 22164215 Free PMC article.
-
Structural analysis of a novel rabbit monoclonal antibody R53 targeting an epitope in HIV-1 gp120 C4 region critical for receptor and co-receptor binding.Emerg Microbes Infect. 2015 Jul;4(7):e44. doi: 10.1038/emi.2015.44. Epub 2015 Jul 15. Emerg Microbes Infect. 2015. PMID: 26251831 Free PMC article.
-
Structural analysis of human and macaque mAbs 2909 and 2.5B: implications for the configuration of the quaternary neutralizing epitope of HIV-1 gp120.Structure. 2011 May 11;19(5):691-9. doi: 10.1016/j.str.2011.02.012. Structure. 2011. PMID: 21565703 Free PMC article.
-
Visualization of retroviral envelope spikes in complex with the V3 loop antibody 447-52D on intact viruses by cryo-electron tomography.J Virol. 2014 Nov;88(21):12265-75. doi: 10.1128/JVI.01596-14. Epub 2014 Aug 13. J Virol. 2014. PMID: 25122783 Free PMC article.
-
Differential induction of anti-V3 crown antibodies with cradle- and ladle-binding modes in response to HIV-1 envelope vaccination.Vaccine. 2017 Mar 7;35(10):1464-1473. doi: 10.1016/j.vaccine.2016.11.107. Epub 2017 Feb 6. Vaccine. 2017. PMID: 28185743 Free PMC article.
References
-
- Abagyan RA, Totrov M, Kuznetsov D. ICM - A new method for protein modeling and design: Applications to docking and structure prediction from the distorted native conformation. Journal of Computational Chemistry. 1994;15:488–506.
-
- Binley JM, Lybarger EA, Crooks ET, Seaman MS, Gray E, Davis KL, Decker JM, Wycuff D, Harris L, Hawkins N, et al. Profiling the specificity of neutralizing antibodies in a large panel of plasmas from patients chronically infected with human immunodeficiency virus type 1 subtypes B and C. J Virol. 2008;82:11651–11668. - PMC - PubMed
-
- Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–921. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

