A Smac-mimetic sensitizes prostate cancer cells to TRAIL-induced apoptosis via modulating both IAPs and NF-kappaB
- PMID: 19895686
- PMCID: PMC2779195
- DOI: 10.1186/1471-2407-9-392
A Smac-mimetic sensitizes prostate cancer cells to TRAIL-induced apoptosis via modulating both IAPs and NF-kappaB
Abstract
Background: Although tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) is a promising agent for human cancer therapy, prostate cancer still remains resistant to TRAIL. Both X-linked inhibitor of apoptosis (XIAP) and nuclear factor-kappaB function as key negative regulators of TRAIL signaling. In this study, we evaluated the effect of SH122, a small molecule mimetic of the second mitochondria-derived activator of caspases (Smac), on TRAIL-induced apoptosis in prostate cancer cells.
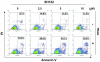
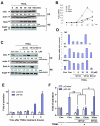
Methods: The potential of Smac-mimetics to bind XIAP or cIAP-1 was examined by pull-down assay. Cytotoxicity of TRAIL and/or Smac-mimetics was determined by a standard cell growth assay. Silencing of XIAP or cIAP-1 was achieved by transient transfection of short hairpin RNA. Apoptosis was detected by Annexin V-PI staining followed by flow cytometry and by Western Blot analysis of caspases, PARP and Bid. NF-kappaB activation was determined by subcellular fractionation, real time RT-PCR and reporter assay.
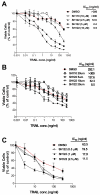
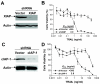
Results: SH122, but not its inactive analog, binds to XIAP and cIAP-1. SH122 significantly sensitized prostate cancer cells to TRAIL-mediated cell death. Moreover, SH122 enhanced TRAIL-induced apoptosis via both the death receptor and the mitochondrial pathway. Knockdown of both XIAP and cIAP-1 sensitized cellular response to TRAIL. XIAP-knockdown attenuated sensitivity of SH122 to TRAIL-induced cytotoxicity, confirming that XIAP is an important target for IAP-inhibitor-mediated TRAIL sensitization. SH122 also suppressed TRAIL-induced NF-kappaB activation by preventing cytosolic IkappaB-alpha degradation and RelA nuclear translocation, as well as by suppressing NF-kappaB target gene expression.
Conclusion: These results demonstrate that SH122 sensitizes human prostate cancer cells to TRAIL-induced apoptosis by mimicking Smac and blocking both IAPs and NF-kappaB. Modulating IAPs may represent a promising approach to overcoming TRAIL-resistance in human prostate cancer with constitutively active NF-kappaB signaling.
Figures



Similar articles
-
X-linked inhibitor of apoptosis (XIAP) blocks Apo2 ligand/tumor necrosis factor-related apoptosis-inducing ligand-mediated apoptosis of prostate cancer cells in the presence of mitochondrial activation: sensitization by overexpression of second mitochondria-derived activator of caspase/direct IAP-binding protein with low pl (Smac/DIABLO).Mol Cancer Ther. 2002 Oct;1(12):1051-8. Mol Cancer Ther. 2002. PMID: 12481428
-
Smac mimetic Birinapant induces apoptosis and enhances TRAIL potency in inflammatory breast cancer cells in an IAP-dependent and TNF-α-independent mechanism.Breast Cancer Res Treat. 2013 Jan;137(2):359-71. doi: 10.1007/s10549-012-2352-6. Epub 2012 Dec 7. Breast Cancer Res Treat. 2013. PMID: 23225169
-
A small molecule Smac-mimic compound induces apoptosis and sensitizes TRAIL- and etoposide-induced apoptosis in breast cancer cells.Oncogene. 2005 Nov 10;24(49):7381-8. doi: 10.1038/sj.onc.1208888. Oncogene. 2005. PMID: 16044155
-
Therapeutic Small Molecules Target Inhibitor of Apoptosis Proteins in Cancers with Deregulation of Extrinsic and Intrinsic Cell Death Pathways.Clin Cancer Res. 2017 Mar 15;23(6):1379-1387. doi: 10.1158/1078-0432.CCR-16-2172. Epub 2016 Dec 30. Clin Cancer Res. 2017. PMID: 28039268 Free PMC article. Review.
-
Regulation of TRAIL-induced apoptosis by ectopic expression of antiapoptotic factors.Vitam Horm. 2004;67:453-83. doi: 10.1016/S0083-6729(04)67023-3. Vitam Horm. 2004. PMID: 15110190 Review.
Cited by
-
Smac mimetics and TRAIL cooperate to induce MLKL-dependent necroptosis in Burkitt's lymphoma cell lines.Neoplasia. 2021 May;23(5):539-550. doi: 10.1016/j.neo.2021.03.003. Epub 2021 May 7. Neoplasia. 2021. PMID: 33971465 Free PMC article.
-
18β-glycyrrhetinic acid potentiates Hsp90 inhibition-induced apoptosis in human epithelial ovarian carcinoma cells via activation of death receptor and mitochondrial pathway.Mol Cell Biochem. 2012 Nov;370(1-2):209-19. doi: 10.1007/s11010-012-1412-x. Epub 2012 Aug 4. Mol Cell Biochem. 2012. PMID: 22865487
-
Smac mimetics LCL161 and GDC-0152 inhibit osteosarcoma growth and metastasis in mice.BMC Cancer. 2019 Sep 14;19(1):924. doi: 10.1186/s12885-019-6103-5. BMC Cancer. 2019. PMID: 31521127 Free PMC article.
-
IAPs on the move: role of inhibitors of apoptosis proteins in cell migration.Cell Death Dis. 2013 Sep 5;4(9):e784. doi: 10.1038/cddis.2013.311. Cell Death Dis. 2013. PMID: 24008728 Free PMC article. Review.
-
Cellular inhibitor of apoptosis 1 (cIAP-1) degradation by caspase 8 during TNF-related apoptosis-inducing ligand (TRAIL)-induced apoptosis.Exp Cell Res. 2011 Jan 1;317(1):107-16. doi: 10.1016/j.yexcr.2010.10.005. Epub 2010 Oct 14. Exp Cell Res. 2011. PMID: 20951133 Free PMC article.
References
-
- Shankar S, Siddiqui I, Srivastava RK. Molecular mechanisms of resveratrol (3,4,5-trihydroxy-trans-stilbene) and its interaction with TNF-related apoptosis inducing ligand (TRAIL) in androgen-insensitive prostate cancer cells. Mol Cell Biochem. 2007;304(1-2):273–285. doi: 10.1007/s11010-007-9510-x. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

