Mitochondrial DNA (mtDNA) biogenesis: visualization and duel incorporation of BrdU and EdU into newly synthesized mtDNA in vitro
- PMID: 19875847
- PMCID: PMC2803709
- DOI: 10.1369/jhc.2009.954701
Mitochondrial DNA (mtDNA) biogenesis: visualization and duel incorporation of BrdU and EdU into newly synthesized mtDNA in vitro
Abstract
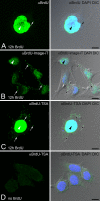
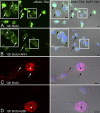
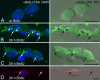
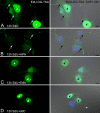
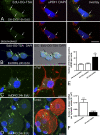
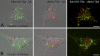
Mitochondria are key regulators of cellular energy and are the focus of a large number of studies examining the regulation of mitochondrial dynamics and biogenesis in healthy and diseased conditions. One approach to monitoring mitochondrial biogenesis is to measure the rate of mitochondrial DNA (mtDNA) replication. We developed a sensitive technique to visualize newly synthesized mtDNA in individual cells to study mtDNA replication within subcellular compartments of neurons. The technique combines the incorporation of 5-bromo-2-deoxyuridine (BrdU) and/or 5-ethynyl-2'-deoxyuridine (EdU) into mtDNA, together with a tyramide signal amplification protocol. Employing this technique, we visualized and measured mtDNA biogenesis in individual cells. The labeling procedure for EdU allows for more comprehensive results by allowing the comparison of its incorporation with other intracellular markers, because it does not require the harsh acid or enzyme digests necessary to recover the BrdU epitope. In addition, the utilization of both BrdU and EdU permits sequential pulse-chase experiments to follow the intracellular localization of mtDNA replication. The ability to quantify mitochondrial biogenesis provides an essential tool for investigating the alterations in mitochondrial dynamics involved in the pathogenesis of multiple cellular disorders, including neuropathies and neurodegenerative diseases.
Figures
Similar articles
-
Visualization of mitochondrial DNA replication in individual cells by EdU signal amplification.J Vis Exp. 2010 Nov 15;(45):2147. doi: 10.3791/2147. J Vis Exp. 2010. PMID: 21113116 Free PMC article.
-
EdU (5-ethynyl-2'-deoxyuridine) labeling of Drosophila mitotic neuroblasts.Cold Spring Harb Protoc. 2010 Jul 1;2010(7):pdb.prot5461. doi: 10.1101/pdb.prot5461. Cold Spring Harb Protoc. 2010. PMID: 20647365
-
In Situ Labeling of Mitochondrial DNA Replication in Drosophila Adult Ovaries by EdU Staining.J Vis Exp. 2016 Oct 15;(116):54516. doi: 10.3791/54516. J Vis Exp. 2016. PMID: 27805603 Free PMC article.
-
Thymidine analogues for tracking DNA synthesis.Molecules. 2011 Sep 15;16(9):7980-93. doi: 10.3390/molecules16097980. Molecules. 2011. PMID: 21921870 Free PMC article. Review.
-
Incorporation of 5-Bromo-2'-deoxyuridine into DNA and Proliferative Behavior of Cerebellar Neuroblasts: All That Glitters Is Not Gold.Cells. 2021 Jun 10;10(6):1453. doi: 10.3390/cells10061453. Cells. 2021. PMID: 34200598 Free PMC article. Review.
Cited by
-
MRE11-dependent instability in mitochondrial DNA fork protection activates a cGAS immune signaling pathway.Sci Adv. 2021 Dec 17;7(51):eabf9441. doi: 10.1126/sciadv.abf9441. Epub 2021 Dec 15. Sci Adv. 2021. PMID: 34910513 Free PMC article.
-
Mitochondrial dynamics regulate growth cone motility, guidance, and neurite growth rate in perinatal retinal ganglion cells in vitro.Invest Ophthalmol Vis Sci. 2012 Oct 30;53(11):7402-11. doi: 10.1167/iovs.12-10298. Invest Ophthalmol Vis Sci. 2012. PMID: 23049086 Free PMC article.
-
DNA Replication: From Radioisotopes to Click Chemistry.Molecules. 2018 Nov 17;23(11):3007. doi: 10.3390/molecules23113007. Molecules. 2018. PMID: 30453631 Free PMC article. Review.
-
Visualization of mitochondrial DNA replication in individual cells by EdU signal amplification.J Vis Exp. 2010 Nov 15;(45):2147. doi: 10.3791/2147. J Vis Exp. 2010. PMID: 21113116 Free PMC article.
-
Downregulation of ALAS1 by nicarbazin treatment underlies the reduced synthesis of protoporphyrin IX in shell gland of laying hens.Sci Rep. 2017 Jul 24;7(1):6253. doi: 10.1038/s41598-017-06527-y. Sci Rep. 2017. PMID: 28740143 Free PMC article.
References
-
- Abou-Sleiman PM, Muqit MM, Wood NW (2006) Expanding insights of mitochondrial dysfunction in Parkinson's disease. Nat Rev Neurosci 7:207–219 - PubMed
-
- Buck SB, Bradford J, Gee KR, Agnew BJ, Clarke ST, Salic A (2008) Detection of S-phase cell cycle progression using 5-ethynyl-2′-deoxyuridine incorporation with click chemistry, an alternative to using 5-bromo-2′-deoxyuridine antibodies. Biotechniques 44:927–929 - PubMed
-
- Cappella P, Gasparri F, Pulici M, Moll J (2008) A novel method based on click chemistry, which overcomes limitations of cell cycle analysis by classical determination of BrdU incorporation, allowing multiplex antibody staining. Cytometry A 73:626–636 - PubMed
-
- Chan DC (2006) Mitochondrial fusion and fission in mammals. Annu Rev Cell Dev Biol 22:79–99 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

