Stimulation of glioma cell motility by expression, proteolysis, and release of the L1 neural cell recognition molecule
- PMID: 19874583
- PMCID: PMC2776596
- DOI: 10.1186/1475-2867-9-27
Stimulation of glioma cell motility by expression, proteolysis, and release of the L1 neural cell recognition molecule
Abstract
Background: Malignant glioma cells are particularly motile and can travel diffusely through the brain parenchyma, apparently without following anatomical structures to guide their migration. The neural adhesion/recognition protein L1 (L1CAM; CD171) has been implicated in contributing to stimulation of motility and metastasis of several non-neural cancer types. We explored the expression and function of L1 protein as a stimulator of glioma cell motility using human high-grade glioma surgical specimens and established rat and human glioma cell lines.
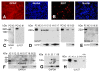
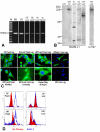
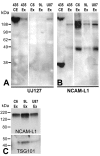
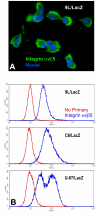
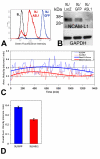
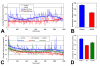
Results: L1 protein expression was found in 17 out of 18 human high-grade glioma surgical specimens by western blotting. L1 mRNA was found to be present in human U-87/LacZ and rat C6 and 9L glioma cell lines. The glioma cell lines were negative for surface full length L1 by flow cytometry and high resolution immunocytochemistry of live cells. However, fixed and permeablized cells exhibited positive staining as numerous intracellular puncta. Western blots of cell line extracts revealed L1 proteolysis into a large soluble ectodomain (~180 kDa) and a smaller transmembrane proteolytic fragment (~32 kDa). Exosomal vesicles released by the glioma cell lines were purified and contained both full-length L1 and the proteolyzed transmembrane fragment. Glioma cell lines expressed L1-binding alphavbeta5 integrin cell surface receptors. Quantitative time-lapse analyses showed that motility was reduced significantly in glioma cell lines by 1) infection with an antisense-L1 retroviral vector and 2) L1 ectodomain-binding antibodies.
Conclusion: Our novel results support a model of autocrine/paracrine stimulation of cell motility in glioma cells by a cleaved L1 ectodomain and/or released exosomal vesicles containing L1. This mechanism could explain the diffuse migratory behavior of high-grade glioma cancer cells within the brain.
Figures
Similar articles
-
Exosomal L1CAM Stimulates Glioblastoma Cell Motility, Proliferation, and Invasiveness.Int J Mol Sci. 2019 Aug 16;20(16):3982. doi: 10.3390/ijms20163982. Int J Mol Sci. 2019. PMID: 31426278 Free PMC article.
-
L1CAM stimulates glioma cell motility and proliferation through the fibroblast growth factor receptor.Clin Exp Metastasis. 2013 Apr;30(4):507-20. doi: 10.1007/s10585-012-9555-4. Epub 2012 Dec 1. Clin Exp Metastasis. 2013. PMID: 23212305
-
L1 stimulation of human glioma cell motility correlates with FAK activation.J Neurooncol. 2011 Oct;105(1):27-44. doi: 10.1007/s11060-011-0557-x. Epub 2011 Mar 4. J Neurooncol. 2011. PMID: 21373966 Free PMC article.
-
Functional Diversity of Neuronal Cell Adhesion and Recognition Molecule L1CAM through Proteolytic Cleavage.Cells. 2022 Sep 30;11(19):3085. doi: 10.3390/cells11193085. Cells. 2022. PMID: 36231047 Free PMC article. Review.
-
Autocrine factors that sustain glioma invasion and paracrine biology in the brain microenvironment.J Natl Cancer Inst. 2007 Nov 7;99(21):1583-93. doi: 10.1093/jnci/djm187. Epub 2007 Oct 30. J Natl Cancer Inst. 2007. PMID: 17971532 Review.
Cited by
-
L1CAM: a major driver for tumor cell invasion and motility.Cell Adh Migr. 2012 Jul-Aug;6(4):374-84. doi: 10.4161/cam.20832. Epub 2012 Jul 1. Cell Adh Migr. 2012. PMID: 22796939 Free PMC article. Review.
-
Exosomal L1CAM Stimulates Glioblastoma Cell Motility, Proliferation, and Invasiveness.Int J Mol Sci. 2019 Aug 16;20(16):3982. doi: 10.3390/ijms20163982. Int J Mol Sci. 2019. PMID: 31426278 Free PMC article.
-
L1CAM stimulates glioma cell motility and proliferation through the fibroblast growth factor receptor.Clin Exp Metastasis. 2013 Apr;30(4):507-20. doi: 10.1007/s10585-012-9555-4. Epub 2012 Dec 1. Clin Exp Metastasis. 2013. PMID: 23212305
-
L1 stimulation of human glioma cell motility correlates with FAK activation.J Neurooncol. 2011 Oct;105(1):27-44. doi: 10.1007/s11060-011-0557-x. Epub 2011 Mar 4. J Neurooncol. 2011. PMID: 21373966 Free PMC article.
-
Comparison of L1 expression and secretion in glioblastoma and neuroblastoma cells.Oncol Lett. 2012 Oct;4(4):812-816. doi: 10.3892/ol.2012.787. Epub 2012 Jul 5. Oncol Lett. 2012. PMID: 23205105 Free PMC article.
References
Grants and funding
LinkOut - more resources
Full Text Sources

