Molecular epidemiology of genogroup II-genotype 4 noroviruses in the United States between 1994 and 2006
- PMID: 19864482
- PMCID: PMC2812295
- DOI: 10.1128/JCM.01622-09
Molecular epidemiology of genogroup II-genotype 4 noroviruses in the United States between 1994 and 2006
Abstract
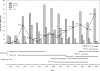
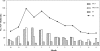
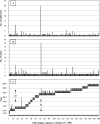
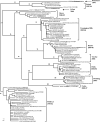
Human noroviruses (NoVs) of genogroup II, genotype 4 (GII.4) are the most common strains detected in outbreaks of acute gastroenteritis worldwide. To gain insight into the epidemiology and genetic variation of GII.4 strains, we analyzed 773 NoV outbreaks reported to the CDC from 1994 to 2006. Of these NoV outbreaks, 629 (81.4%) were caused by GII viruses and 342 (44.2%) were caused by GII.4 strains. The proportion of GII.4 outbreaks increased from 5% in 1994 to 85% in 2006, but distinct annual differences were noted, including sharp increases in 1996, 2003, and 2006 each associated with newly emerging GII.4 strains. Sequence analysis of the full-length VP1 gene of GII.4 strains identified in this study and from GenBank segregated these viruses into at least 9 distinct subclusters which had 1.3 to 3.2% amino acid variation between strains in different subclusters. We propose that GII.4 subclusters be defined as having >5% sequence variation between strains. Our data confirm other studies on the rapid emergence and displacement of highly virulent GII.4 strains.
Figures


Similar articles
-
Genetic diversity and epidemiology of Genogroup II noroviruses in children with acute sporadic gastroenteritis in Shanghai, China, 2012-2017.BMC Infect Dis. 2019 Aug 22;19(1):736. doi: 10.1186/s12879-019-4360-1. BMC Infect Dis. 2019. PMID: 31438883 Free PMC article.
-
Molecular epidemiology of noroviruses associated with sporadic gastroenteritis in children in Novosibirsk, Russia, 2003-2012.J Med Virol. 2015 May;87(5):740-53. doi: 10.1002/jmv.24068. Epub 2015 Feb 18. J Med Virol. 2015. PMID: 25693507
-
Molecular and clinical epidemiology of norovirus outbreaks in Spain during the emergence of GII.4 2012 variant.J Clin Virol. 2014 Jun;60(2):96-104. doi: 10.1016/j.jcv.2014.03.013. Epub 2014 Mar 27. J Clin Virol. 2014. PMID: 24746342
-
Molecular epidemiology and genotype distributions of noroviruses and sapoviruses in Thailand 2000-2016: A review.J Med Virol. 2018 Apr;90(4):617-624. doi: 10.1002/jmv.25019. Epub 2018 Jan 23. J Med Virol. 2018. PMID: 29315631 Review.
-
Molecular epidemiology of noroviruses associated with acute sporadic gastroenteritis in children: global distribution of genogroups, genotypes and GII.4 variants.J Clin Virol. 2013 Mar;56(3):185-93. doi: 10.1016/j.jcv.2012.11.011. Epub 2012 Dec 5. J Clin Virol. 2013. PMID: 23218993 Review.
Cited by
-
Norovirus Illnesses in Children and Adolescents.Infect Dis Clin North Am. 2018 Mar;32(1):103-118. doi: 10.1016/j.idc.2017.11.004. Infect Dis Clin North Am. 2018. PMID: 29406972 Free PMC article. Review.
-
Near Real-Time Surveillance of U.S. Norovirus Outbreaks by the Norovirus Sentinel Testing and Tracking Network - United States, August 2009-July 2015.MMWR Morb Mortal Wkly Rep. 2017 Feb 24;66(7):185-189. doi: 10.15585/mmwr.mm6607a1. MMWR Morb Mortal Wkly Rep. 2017. PMID: 28231235 Free PMC article.
-
A systematic review and meta-analysis of the prevalence of norovirus in cases of gastroenteritis in developing countries.Medicine (Baltimore). 2017 Oct;96(40):e8139. doi: 10.1097/MD.0000000000008139. Medicine (Baltimore). 2017. PMID: 28984764 Free PMC article. Review.
-
Proposal for a unified norovirus nomenclature and genotyping.Arch Virol. 2013 Oct;158(10):2059-68. doi: 10.1007/s00705-013-1708-5. Epub 2013 Apr 25. Arch Virol. 2013. PMID: 23615870 Free PMC article.
-
Development of a Simple, Fast, and Cost-Effective Nanobased Immunoassay Method for Detecting Norovirus in Food Samples.ACS Omega. 2020 May 19;5(21):12162-12165. doi: 10.1021/acsomega.0c00502. eCollection 2020 Jun 2. ACS Omega. 2020. PMID: 32548397 Free PMC article.
References
-
- Ando, T., M. N. Mulders, D. C. Lewis, M. K. Estes, S. S. Monroe, and R. I. Glass. 1994. Comparison of the polymerase region of small round structured virus strains previously classified in three antigenic types by solid-phase immune electron microscopy. Arch. Virol. 135:217-226. - PubMed
-
- Blanton, L. H., S. M. Adams, R. S. Beard, G. Wei, S. N. Bulens, M. A. Widdowson, R. I. Glass, and S. S. Monroe. 2006. Molecular and epidemiologic trends of caliciviruses associated with outbreaks of acute gastroenteritis in the United States, 2000-2004. J. Infect. Dis. 193:413-421. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

