Firefly luciferase: an adenylate-forming enzyme for multicatalytic functions
- PMID: 19859663
- PMCID: PMC11115821
- DOI: 10.1007/s00018-009-0170-8
Firefly luciferase: an adenylate-forming enzyme for multicatalytic functions
Abstract
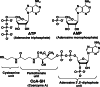
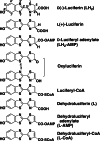
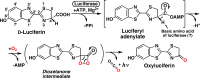
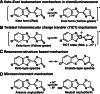
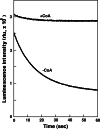
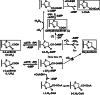
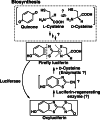
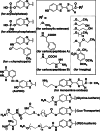
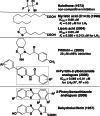
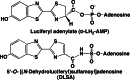
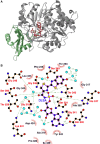
Firefly luciferase is a member of the acyl-adenylate/thioester-forming superfamily of enzymes and catalyzes the oxidation of firefly luciferin with molecular oxygen to emit light. Knowledge of the luminescence mechanism catalyzed by firefly luciferase has been gathered, leading to the discovery of a novel catalytic function of luciferase. Recently, we demonstrated that firefly luciferase has a catalytic function of fatty acyl-CoA synthesis from fatty acids in the presence of ATP, Mg(2+) and coenzyme A. Based on identification of fatty acyl-CoA genes in firefly, Drosophila, and non-luminous click beetles, we then proposed that the evolutionary origin of firefly luciferase is a fatty acyl-CoA synthetase in insects. Further, we succeeded in converting the fatty acyl-CoA synthetase of non-luminous insects into functional luciferase showing luminescence activity by site-directed mutagenesis.
Figures

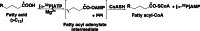


Similar articles
-
Functional conversion of fatty acyl-CoA synthetase to firefly luciferase by site-directed mutagenesis: a key substitution responsible for luminescence activity.FEBS Lett. 2009 Jun 18;583(12):2004-8. doi: 10.1016/j.febslet.2009.05.018. Epub 2009 May 18. FEBS Lett. 2009. PMID: 19450587
-
Catalytic properties of domain-exchanged chimeric proteins between firefly luciferase and Drosophila fatty Acyl-CoA synthetase CG6178.Biosci Biotechnol Biochem. 2006 Nov;70(11):2739-44. doi: 10.1271/bbb.60364. Epub 2006 Nov 7. Biosci Biotechnol Biochem. 2006. PMID: 17090919
-
Characterization of luciferases and its paralogue in the Panamanian luminous click beetle Pyrophorus angustus: a click beetle luciferase lacks the fatty acyl-CoA synthetic activity.Gene. 2010 Feb 15;452(1):1-6. doi: 10.1016/j.gene.2009.12.001. Epub 2009 Dec 11. Gene. 2010. PMID: 20004235
-
Molecular enigma of multicolor bioluminescence of firefly luciferase.Cell Mol Life Sci. 2011 Apr;68(7):1167-82. doi: 10.1007/s00018-010-0607-0. Epub 2010 Dec 28. Cell Mol Life Sci. 2011. PMID: 21188462 Free PMC article. Review.
-
Enzymatic promiscuity and the evolution of bioluminescence.FEBS J. 2020 Apr;287(7):1369-1380. doi: 10.1111/febs.15176. Epub 2019 Dec 27. FEBS J. 2020. PMID: 31828943 Free PMC article. Review.
Cited by
-
Aristolochic acid I exposure decreases oocyte quality.Front Cell Dev Biol. 2022 Aug 11;10:838992. doi: 10.3389/fcell.2022.838992. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 36036003 Free PMC article.
-
In-depth Characterization of Firefly Luciferase as a Reporter of Circadian Gene Expression in Mammalian Cells.J Biol Rhythms. 2016 Dec;31(6):540-550. doi: 10.1177/0748730416668898. Epub 2016 Oct 10. J Biol Rhythms. 2016. PMID: 28112045 Free PMC article.
-
Rapid obtention of stable, bioluminescent tumor cell lines using a tCD2-luciferase chimeric construct.BMC Biotechnol. 2011 Mar 24;11:26. doi: 10.1186/1472-6750-11-26. BMC Biotechnol. 2011. PMID: 21435248 Free PMC article.
-
Comparative RNA seq analysis of the New Zealand glowworm Arachnocampa luminosa reveals bioluminescence-related genes.BMC Genomics. 2015 Oct 21;16:825. doi: 10.1186/s12864-015-2006-2. BMC Genomics. 2015. PMID: 26486607 Free PMC article.
-
A luciferin analogue generating near-infrared bioluminescence achieves highly sensitive deep-tissue imaging.Nat Commun. 2016 Jun 14;7:11856. doi: 10.1038/ncomms11856. Nat Commun. 2016. PMID: 27297211 Free PMC article.
References
-
- Harvey EN. Bioluminescence. New York: Academic; 1952.
-
- Haneda Y, Johnson FH, editors. Bioluminescence in progress. New Jersey: Princeton University Press; 1966.
-
- Herring PJ, editor. Bioluminescence in action. New York: Academic; 1978.
-
- Campbell AK. Chemiluminescence: principle and applications in biology and medicine. New York: VCH; 1988.
-
- Shimomura O. Bioluminescence: chemical principles and methods. Singapore: World Scientific; 2006.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

