Epigenetic activation of unintegrated HIV-1 genomes by gut-associated short chain fatty acids and its implications for HIV infection
- PMID: 19843699
- PMCID: PMC2773968
- DOI: 10.1073/pnas.0905859106
Epigenetic activation of unintegrated HIV-1 genomes by gut-associated short chain fatty acids and its implications for HIV infection
Abstract
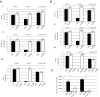
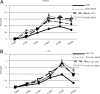
Integration of HIV-1 linear DNA into the host chromatin is an essential step in the viral life cycle. However, the majority of reverse-transcribed, nuclear-imported viral genomes remain episomal, either as linear or circular DNA. To date, these nonintegrated viral genomes are largely considered "dead-end products" of reverse transcription. Indeed, limited gene expression from nonintegrated HIV-1 has been reported, although the mechanism that renders nonintegrating HIV-1 genomes incapable of supporting efficient viral replication has not been fully elucidated. Here, we demonstrate that nonintegrating HIV-1 and HIV-1-based vector genomes are organized into chromatin structures and enriched with histone modifications typical of transcriptionally silenced chromatin. Gene expression and replication of nonintegrating HIV-1 was notably increased in vitro upon exposure to histone deacetylase inhibitors (HDACi) in the form of various short-chain fatty acids (SCFAs) known to be endogenously produced by normal microbial-gut flora. Furthermore, we demonstrated genetic and functional crosstalk between episomal and integrated vector/viral genomes, resulting in recombination between integrated and nonintegrated HIV-1, as well as mobilization of episomal vector genomes by productive viral particles encoded by integrated viral genomes. Finally, we propose a mechanism describing the role of episomal HIV-1 forms in the viral life cycle in a SCFA-rich gut environment.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Reversal of Epigenetic Silencing Allows Robust HIV-1 Replication in the Absence of Integrase Function.mBio. 2020 Jun 2;11(3):e01038-20. doi: 10.1128/mBio.01038-20. mBio. 2020. PMID: 32487757 Free PMC article.
-
Tax Induces the Recruitment of NF-κB to Unintegrated HIV-1 DNA To Rescue Viral Gene Expression and Replication.J Virol. 2021 Jun 10;95(13):e0028521. doi: 10.1128/JVI.00285-21. Epub 2021 Jun 10. J Virol. 2021. PMID: 33883218 Free PMC article.
-
Epigenetic Metabolite Acetate Inhibits Class I/II Histone Deacetylases, Promotes Histone Acetylation, and Increases HIV-1 Integration in CD4+ T Cells.J Virol. 2017 Jul 27;91(16):e01943-16. doi: 10.1128/JVI.01943-16. Print 2017 Aug 15. J Virol. 2017. PMID: 28539453 Free PMC article.
-
Epigenetic regulation of HIV-1 transcription.Epigenomics. 2011 Aug;3(4):487-502. doi: 10.2217/epi.11.61. Epigenomics. 2011. PMID: 22126207 Review.
-
Chromatin-associated regulation of HIV-1 transcription: implications for the development of therapeutic strategies.Subcell Biochem. 2007;41:371-96. Subcell Biochem. 2007. PMID: 17484137 Review.
Cited by
-
The gut microbiota and its metabolite butyrate shape metabolism and antiviral immunity along the gut-lung axis in the chicken.Commun Biol. 2024 Sep 20;7(1):1185. doi: 10.1038/s42003-024-06815-0. Commun Biol. 2024. PMID: 39300162 Free PMC article.
-
Integrase inhibitor reversal dynamics indicate unintegrated HIV-1 dna initiate de novo integration.Retrovirology. 2015 Mar 12;12:24. doi: 10.1186/s12977-015-0153-9. Retrovirology. 2015. PMID: 25808736 Free PMC article.
-
Impact of periodontal intervention on local inflammation, periodontitis, and HIV outcomes.Oral Dis. 2016 Apr;22 Suppl 1(Suppl 1):87-97. doi: 10.1111/odi.12419. Oral Dis. 2016. PMID: 27109277 Free PMC article.
-
Tat IRES modulator of tat mRNA (TIM-TAM): a conserved RNA structure that controls Tat expression and acts as a switch for HIV productive and latent infection.Nucleic Acids Res. 2020 Mar 18;48(5):2643-2660. doi: 10.1093/nar/gkz1181. Nucleic Acids Res. 2020. PMID: 31875221 Free PMC article.
-
Change of histone H3 lysine 14 acetylation stoichiometry in human monocyte derived macrophages as determined by MS-based absolute targeted quantitative proteomic approach: HIV infection and methamphetamine exposure.Clin Proteomics. 2023 Oct 25;20(1):48. doi: 10.1186/s12014-023-09438-5. Clin Proteomics. 2023. PMID: 37880620 Free PMC article.
References
-
- Chun T, et al. Quantification of latent tissue reservoirs and total body viral load in HIV-1 infection. Nature. 1997;387:183–188. - PubMed
-
- Gillim-Ross L, Cara A, Klotman ME. HIV-1 extrachromosomal 2-LTRs circular DNA is long-lived in human macrophages. Viral Immunol. 2005;18:190–196. - PubMed
-
- Cara A, Guarnaccia F, Reitz MS, Gallo RC, Lori F. Self-limiting, cell type-dependent replication of an integrase-defective human immunodeficiency virus type 1 in human primary macrophages but not T lymphocytes. Virology. 1995;208:242–248. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

