Recurrent microdeletions at 15q11.2 and 16p13.11 predispose to idiopathic generalized epilepsies
- PMID: 19843651
- PMCID: PMC2801323
- DOI: 10.1093/brain/awp262
Recurrent microdeletions at 15q11.2 and 16p13.11 predispose to idiopathic generalized epilepsies
Abstract
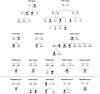
Idiopathic generalized epilepsies account for 30% of all epilepsies. Despite a predominant genetic aetiology, the genetic factors predisposing to idiopathic generalized epilepsies remain elusive. Studies of structural genomic variations have revealed a significant excess of recurrent microdeletions at 1q21.1, 15q11.2, 15q13.3, 16p11.2, 16p13.11 and 22q11.2 in various neuropsychiatric disorders including autism, intellectual disability and schizophrenia. Microdeletions at 15q13.3 have recently been shown to constitute a strong genetic risk factor for common idiopathic generalized epilepsy syndromes, implicating that other recurrent microdeletions may also be involved in epileptogenesis. This study aimed to investigate the impact of five microdeletions at the genomic hotspot regions 1q21.1, 15q11.2, 16p11.2, 16p13.11 and 22q11.2 on the genetic risk to common idiopathic generalized epilepsy syndromes. The candidate microdeletions were assessed by high-density single nucleotide polymorphism arrays in 1234 patients with idiopathic generalized epilepsy from North-western Europe and 3022 controls from the German population. Microdeletions were validated by quantitative polymerase chain reaction and their breakpoints refined by array comparative genomic hybridization. In total, 22 patients with idiopathic generalized epilepsy (1.8%) carried one of the five novel microdeletions compared with nine controls (0.3%) (odds ratio = 6.1; 95% confidence interval 2.8-13.2; chi(2) = 26.7; 1 degree of freedom; P = 2.4 x 10(-7)). Microdeletions were observed at 1q21.1 [Idiopathic generalized epilepsy (IGE)/control: 1/1], 15q11.2 (IGE/control: 12/6), 16p11.2 IGE/control: 1/0, 16p13.11 (IGE/control: 6/2) and 22q11.2 (IGE/control: 2/0). Significant associations with IGEs were found for the microdeletions at 15q11.2 (odds ratio = 4.9; 95% confidence interval 1.8-13.2; P = 4.2 x 10(-4)) and 16p13.11 (odds ratio = 7.4; 95% confidence interval 1.3-74.7; P = 0.009). Including nine patients with idiopathic generalized epilepsy in this cohort with known 15q13.3 microdeletions (IGE/control: 9/0), parental transmission could be examined in 14 families. While 10 microdeletions were inherited (seven maternal and three paternal transmissions), four microdeletions occurred de novo at 15q13.3 (n = 1), 16p13.11 (n = 2) and 22q11.2 (n = 1). Eight of the transmitting parents were clinically unaffected, suggesting that the microdeletion itself is not sufficient to cause the epilepsy phenotype. Although the microdeletions investigated are individually rare (<1%) in patients with idiopathic generalized epilepsy, they collectively seem to account for a significant fraction of the genetic variance in common idiopathic generalized epilepsy syndromes. The present results indicate an involvement of microdeletions at 15q11.2 and 16p13.11 in epileptogenesis and strengthen the evidence that recurrent microdeletions at 15q11.2, 15q13.3 and 16p13.11 confer a pleiotropic susceptibility effect to a broad range of neuropsychiatric disorders.
Figures


Comment in
-
Copy number variants--an unexpected risk factor for the idiopathic generalized epilepsies.Brain. 2010 Jan;133(Pt 1):7-8. doi: 10.1093/brain/awp332. Brain. 2010. PMID: 20047903 Review. No abstract available.
Similar articles
-
Iterative phenotyping of 15q11.2, 15q13.3 and 16p13.11 microdeletion carriers in pediatric epilepsies.Epilepsy Res. 2014 Jan;108(1):109-16. doi: 10.1016/j.eplepsyres.2013.10.001. Epub 2013 Oct 26. Epilepsy Res. 2014. PMID: 24246141
-
[Submicroscopic chromosomal changes predispose to generalised epilepsy].Ugeskr Laeger. 2011 Apr 18;173(16-17):1201-4. Ugeskr Laeger. 2011. PMID: 21501562 Danish.
-
15q13.3 microdeletions in a prospectively recruited cohort of patients with idiopathic generalized epilepsy in Bulgaria.Epilepsy Res. 2013 May;104(3):241-5. doi: 10.1016/j.eplepsyres.2012.10.013. Epub 2013 Jan 23. Epilepsy Res. 2013. PMID: 23352738
-
Copy number variants are frequent in genetic generalized epilepsy with intellectual disability.Neurology. 2013 Oct 22;81(17):1507-14. doi: 10.1212/WNL.0b013e3182a95829. Epub 2013 Sep 25. Neurology. 2013. PMID: 24068782 Free PMC article. Review.
-
Epilepsy and the new cytogenetics.Epilepsia. 2011 Mar;52(3):423-32. doi: 10.1111/j.1528-1167.2010.02932.x. Epub 2011 Jan 26. Epilepsia. 2011. PMID: 21269290 Free PMC article. Review.
Cited by
-
Analysis of rare copy number variation in absence epilepsies.Neurol Genet. 2016 Mar 22;2(2):e56. doi: 10.1212/NXG.0000000000000056. eCollection 2016 Apr. Neurol Genet. 2016. PMID: 27123475 Free PMC article.
-
The genetic variability and commonality of neurodevelopmental disease.Am J Med Genet C Semin Med Genet. 2012 May 15;160C(2):118-29. doi: 10.1002/ajmg.c.31327. Epub 2012 Apr 12. Am J Med Genet C Semin Med Genet. 2012. PMID: 22499536 Free PMC article. Review.
-
Familial cosegregation of rare genetic variants with disease in complex disorders.Eur J Hum Genet. 2013 Apr;21(4):444-50. doi: 10.1038/ejhg.2012.194. Epub 2012 Sep 26. Eur J Hum Genet. 2013. PMID: 23010752 Free PMC article.
-
Distribution of disease-associated copy number variants across distinct disorders of cognitive development.J Am Acad Child Adolesc Psychiatry. 2013 Apr;52(4):414-430.e14. doi: 10.1016/j.jaac.2013.01.003. J Am Acad Child Adolesc Psychiatry. 2013. PMID: 23582872 Free PMC article. Review.
-
Rare copy number variants are an important cause of epileptic encephalopathies.Ann Neurol. 2011 Dec;70(6):974-85. doi: 10.1002/ana.22645. Ann Neurol. 2011. PMID: 22190369 Free PMC article.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

