AMPK regulates the circadian clock by cryptochrome phosphorylation and degradation
- PMID: 19833968
- PMCID: PMC2819106
- DOI: 10.1126/science.1172156
AMPK regulates the circadian clock by cryptochrome phosphorylation and degradation
Abstract
Circadian clocks coordinate behavioral and physiological processes with daily light-dark cycles by driving rhythmic transcription of thousands of genes. Whereas the master clock in the brain is set by light, pacemakers in peripheral organs, such as the liver, are reset by food availability, although the setting, or "entrainment," mechanisms remain mysterious. Studying mouse fibroblasts, we demonstrated that the nutrient-responsive adenosine monophosphate-activated protein kinase (AMPK) phosphorylates and destabilizes the clock component cryptochrome 1 (CRY1). In mouse livers, AMPK activity and nuclear localization were rhythmic and inversely correlated with CRY1 nuclear protein abundance. Stimulation of AMPK destabilized cryptochromes and altered circadian rhythms, and mice in which the AMPK pathway was genetically disrupted showed alterations in peripheral clocks. Thus, phosphorylation by AMPK enables cryptochrome to transduce nutrient signals to circadian clocks in mammalian peripheral organs.
Figures
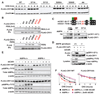
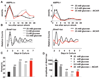

Comment in
-
Physiology. Feeding the clock.Science. 2009 Oct 16;326(5951):378-9. doi: 10.1126/science.1181278. Science. 2009. PMID: 19833950 No abstract available.
Similar articles
-
Relationship between AMPK and the transcriptional balance of clock-related genes in skeletal muscle.Am J Physiol Endocrinol Metab. 2008 Nov;295(5):E1032-7. doi: 10.1152/ajpendo.90510.2008. Epub 2008 Aug 26. Am J Physiol Endocrinol Metab. 2008. PMID: 18728219
-
Functional evolution of the photolyase/cryptochrome protein family: importance of the C terminus of mammalian CRY1 for circadian core oscillator performance.Mol Cell Biol. 2006 Mar;26(5):1743-53. doi: 10.1128/MCB.26.5.1743-1753.2006. Mol Cell Biol. 2006. PMID: 16478995 Free PMC article.
-
AMPK at the crossroads of circadian clocks and metabolism.Mol Cell Endocrinol. 2013 Feb 25;366(2):163-9. doi: 10.1016/j.mce.2012.06.017. Epub 2012 Jun 28. Mol Cell Endocrinol. 2013. PMID: 22750052 Free PMC article. Review.
-
Cycling of CRYPTOCHROME proteins is not necessary for circadian-clock function in mammalian fibroblasts.Curr Biol. 2007 Jul 3;17(13):1091-100. doi: 10.1016/j.cub.2007.05.048. Epub 2007 Jun 21. Curr Biol. 2007. PMID: 17583506 Free PMC article.
-
Genetics and neurobiology of circadian clocks in mammals.Cold Spring Harb Symp Quant Biol. 2007;72:251-259. doi: 10.1101/sqb.2007.72.052. Cold Spring Harb Symp Quant Biol. 2007. PMID: 18419282 Free PMC article. Review.
Cited by
-
Circadian timekeeping and output mechanisms in animals.Curr Opin Neurobiol. 2013 Oct;23(5):724-31. doi: 10.1016/j.conb.2013.02.018. Epub 2013 May 31. Curr Opin Neurobiol. 2013. PMID: 23731779 Free PMC article. Review.
-
USP7 and TDP-43: Pleiotropic Regulation of Cryptochrome Protein Stability Paces the Oscillation of the Mammalian Circadian Clock.PLoS One. 2016 Apr 28;11(4):e0154263. doi: 10.1371/journal.pone.0154263. eCollection 2016. PLoS One. 2016. PMID: 27123980 Free PMC article.
-
Targeting of the circadian clock via CK1δ/ε to improve glucose homeostasis in obesity.Sci Rep. 2016 Jul 21;6:29983. doi: 10.1038/srep29983. Sci Rep. 2016. PMID: 27439882 Free PMC article.
-
Interplay between Metabolism and Epigenetics: A Nuclear Adaptation to Environmental Changes.Mol Cell. 2016 Jun 2;62(5):695-711. doi: 10.1016/j.molcel.2016.05.029. Mol Cell. 2016. PMID: 27259202 Free PMC article. Review.
-
New insights into non-transcriptional regulation of mammalian core clock proteins.J Cell Sci. 2020 Sep 15;133(18):jcs241174. doi: 10.1242/jcs.241174. J Cell Sci. 2020. PMID: 32934011 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
- R01 DK080425-03/DK/NIDDK NIH HHS/United States
- R01 DK080425/DK/NIDDK NIH HHS/United States
- DK062434/DK/NIDDK NIH HHS/United States
- CA104838/CA/NCI NIH HHS/United States
- U19 DK062434/DK/NIDDK NIH HHS/United States
- P01 CA104838-05S1/CA/NCI NIH HHS/United States
- R01 EY016807-03/EY/NEI NIH HHS/United States
- P01 CA104838/CA/NCI NIH HHS/United States
- R37 DK057978/DK/NIDDK NIH HHS/United States
- EY016807/EY/NEI NIH HHS/United States
- R01 EY016807/EY/NEI NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- R37 DK057978-31/DK/NIDDK NIH HHS/United States
- U19 DK062434-08S19002/DK/NIDDK NIH HHS/United States
- DK057978/DK/NIDDK NIH HHS/United States
- T32 HL007439/HL/NHLBI NIH HHS/United States
- T32-HL07439-27/HL/NHLBI NIH HHS/United States
- T32 HL007439-27/HL/NHLBI NIH HHS/United States
- DK080425/DK/NIDDK NIH HHS/United States
- P30 CA014195/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

