Regulation by salt of vacuolar H+-ATPase and H+-pyrophosphatase activities and Na+/H+ exchange
- PMID: 19820346
- PMCID: PMC2801382
- DOI: 10.4161/psb.4.8.9236
Regulation by salt of vacuolar H+-ATPase and H+-pyrophosphatase activities and Na+/H+ exchange
Abstract
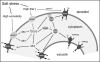
Over the last decades several efforts have been carried out to determine the mechanisms of salt homeostasis in plants and, more recently, to identify genes implicated in salt tolerance, with some plants being successfully genetically engineered to improve resistance to salt. It is well established that the efficient exclusion of Na(+) excess from the cytoplasm and vacuolar Na(+) accumulation are the most important steps towards the maintenance of ion homeostasis inside the cell. Therefore, the vacuole of plant cells plays a pivotal role in the storage of salt. After the identification of the vacuolar Na(+)/H(+) antiporter Nhx1 in Saccharomyces cerevisiae, the first plant Na(+)/H(+) antiporter, AtNHX1, was isolated from Arabidopsis and its overexpression resulted in plants exhibiting increased salt tolerance. Also, the identification of the plasma membrane Na(+)/H(+) exchanger SOS1 and how it is regulated by a protein kinase SOS2 and a calcium binding protein SOS3 were great achievements in the understanding of plant salt resistance. Both tonoplast and plasma membrane antiporters exclude Na+ from the cytosol driven by the proton-motive force generated by the plasma membrane H(+)-ATPase and by the vacuolar membrane H(+)-ATPase and H(+)-pyrophosphatase and it has been shown that the activity of these proteins responds to salinity. In this review we focus on the transcriptional and post-transcriptional regulation by salt of tonoplast proton pumps and Na(+)/H(+) exchangers and on the signalling pathways involved in salt sensing.
Figures



Similar articles
-
Cloning and characterization of a wheat vacuolar cation/proton antiporter and pyrophosphatase proton pump.Plant Physiol Biochem. 2005 Apr;43(4):347-54. doi: 10.1016/j.plaphy.2005.02.010. Epub 2005 Mar 17. Plant Physiol Biochem. 2005. PMID: 15907686
-
Activity of tonoplast proton pumps and Na+/H+ exchange in potato cell cultures is modulated by salt.J Exp Bot. 2009;60(4):1363-74. doi: 10.1093/jxb/erp011. Epub 2009 Feb 12. J Exp Bot. 2009. PMID: 19213810
-
High V-PPase activity is beneficial under high salt loads, but detrimental without salinity.New Phytol. 2018 Sep;219(4):1421-1432. doi: 10.1111/nph.15280. Epub 2018 Jun 25. New Phytol. 2018. PMID: 29938800 Free PMC article.
-
Post-translational regulation of the membrane transporters contributing to salt tolerance in plants.Funct Plant Biol. 2021 Nov;48(12):1199-1212. doi: 10.1071/FP21153. Funct Plant Biol. 2021. PMID: 34665998 Review.
-
Ion homeostasis during salt stress in plants.Curr Opin Cell Biol. 2001 Aug;13(4):399-404. doi: 10.1016/s0955-0674(00)00227-1. Curr Opin Cell Biol. 2001. PMID: 11454443 Review.
Cited by
-
Ion Changes and Signaling under Salt Stress in Wheat and Other Important Crops.Plants (Basel). 2023 Dec 22;13(1):46. doi: 10.3390/plants13010046. Plants (Basel). 2023. PMID: 38202354 Free PMC article. Review.
-
Temporal and spatial changes in ion homeostasis, antioxidant defense and accumulation of flavonoids and glycolipid in a halophyte Sesuvium portulacastrum (L.) L.PLoS One. 2018 Apr 11;13(4):e0193394. doi: 10.1371/journal.pone.0193394. eCollection 2018. PLoS One. 2018. PMID: 29641593 Free PMC article.
-
Identification and computational annotation of genes differentially expressed in pulp development of Cocos nucifera L. by suppression subtractive hybridization.BMC Plant Biol. 2014 Aug 2;14:205. doi: 10.1186/s12870-014-0205-7. BMC Plant Biol. 2014. PMID: 25084812 Free PMC article.
-
Identification of Proteins Modulated in the Date Palm Stem Infested with Red Palm Weevil (Rhynchophorus ferrugineus Oliv.) Using Two Dimensional Differential Gel Electrophoresis and Mass Spectrometry.Int J Mol Sci. 2015 Aug 17;16(8):19326-46. doi: 10.3390/ijms160819326. Int J Mol Sci. 2015. PMID: 26287180 Free PMC article.
-
Vacuolar H+-ATPase works in parallel with the HOG pathway to adapt Saccharomyces cerevisiae cells to osmotic stress.Eukaryot Cell. 2012 Mar;11(3):282-91. doi: 10.1128/EC.05198-11. Epub 2011 Dec 30. Eukaryot Cell. 2012. PMID: 22210831 Free PMC article.
References
-
- Rengasamy P. World salinization with emphasis on Australia. J Exp Bot. 2006;57:1017–1023. - PubMed
-
- Sahi C, Singh A, Blumwald E, Grover A. Beyond osmolytes and transporters: novel plant salt-stress tolerance-related genes from transcriptional profiling data. Physiol Plantarum. 2006;127:1–9.
-
- Blumwald E, Aharon GS, Apse MP. Sodium transport in plant cells. Biochim Biophys Acta. 2000;1465:140–151. - PubMed
-
- Higinbotham N. Electropotentials of plant cells. Annu Rev Plant Physiol. 1973;24:25–46.
-
- Flowers TJ. Improving crop salt tolerance. J Exp Bot. 2004;55:307–319. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous
