Clonality of mouse and human cardiomyogenesis in vivo
- PMID: 19805158
- PMCID: PMC2761310
- DOI: 10.1073/pnas.0903089106
Clonality of mouse and human cardiomyogenesis in vivo
Abstract
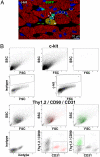
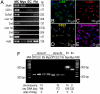
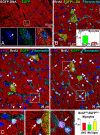
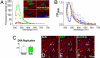
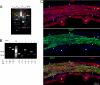
An analysis of the clonality of cardiac progenitor cells (CPCs) and myocyte turnover in vivo requires genetic tagging of the undifferentiated cells so that the clonal marker of individual mother cells is traced in the specialized progeny. CPC niches in the atria and apex of the mouse heart were infected with a lentivirus carrying EGFP, and the destiny of the tagged cells was determined 1-5 months later. A common integration site was identified in isolated CPCs, cardiomyocytes, endothelial cells (ECs), and fibroblasts, documenting CPC self-renewal and multipotentiality and the clonal origin of the differentiated cell populations. Subsequently, the degree of EGFP-lentiviral infection of CPCs was evaluated 2-4 days after injection, and the number of myocytes expressing the reporter gene was measured 6 months later. A BrdU pulse-chasing protocol was also introduced as an additional assay for the analysis of myocyte turnover. Over a period of 6 months, each EGFP-positive CPC divided approximately eight times generating 230 cardiomyocytes; this value was consistent with the number of newly formed cells labeled by BrdU. To determine whether, human CPCs (hCPCs) are self-renewing and multipotent, these cells were transduced with the EGFP-lentivirus and injected after acute myocardial infarction in immunosuppressed rats. hCPCs, myocytes, ECs, and fibroblasts collected from the regenerated myocardium showed common viral integration sites in the human genome. Thus, our results indicate that the adult heart contains a pool of resident stem cells that regulate cardiac homeostasis and repair.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Adult cardiac stem cells are multipotent and support myocardial regeneration.Cell. 2003 Sep 19;114(6):763-76. doi: 10.1016/s0092-8674(03)00687-1. Cell. 2003. PMID: 14505575
-
Isolation and characterization of a Sca-1+/CD31- progenitor cell lineage derived from mouse heart tissue.BMC Biotechnol. 2014 Aug 9;14:75. doi: 10.1186/1472-6750-14-75. BMC Biotechnol. 2014. PMID: 25106452 Free PMC article.
-
[Construction of rat bdnf gene lentiviral vector and its expression in mesenchymal stem cells].Sheng Wu Gong Cheng Xue Bao. 2007 Mar;23(2):235-40. doi: 10.1016/s1872-2075(07)60020-x. Sheng Wu Gong Cheng Xue Bao. 2007. PMID: 17460894 Chinese.
-
Strategies of conditional gene expression in myocardium: an overview.Methods Mol Med. 2005;112:109-54. doi: 10.1007/978-1-59259-879-3_8. Methods Mol Med. 2005. PMID: 16010014 Review.
-
Resident human cardiac stem cells: role in cardiac cellular homeostasis and potential for myocardial regeneration.Nat Clin Pract Cardiovasc Med. 2006 Mar;3 Suppl 1:S8-13. doi: 10.1038/ncpcardio0409. Nat Clin Pract Cardiovasc Med. 2006. PMID: 16501638 Review.
Cited by
-
Single-cell analysis of the fate of c-kit-positive bone marrow cells.NPJ Regen Med. 2017 Oct 16;2:27. doi: 10.1038/s41536-017-0032-1. eCollection 2017. NPJ Regen Med. 2017. PMID: 29302361 Free PMC article.
-
Cardiomyocyte proliferation vs progenitor cells in myocardial regeneration: The debate continues.Glob Cardiol Sci Pract. 2013 Nov 1;2013(3):303-15. doi: 10.5339/gcsp.2013.37. eCollection 2013. Glob Cardiol Sci Pract. 2013. PMID: 24689031 Free PMC article. Review.
-
Human cardiac stem cell differentiation is regulated by a mircrine mechanism.Circulation. 2011 Mar 29;123(12):1287-96. doi: 10.1161/CIRCULATIONAHA.110.982918. Epub 2011 Mar 14. Circulation. 2011. Retraction in: Circulation. 2019 Feb 12;139(7):e38. doi: 10.1161/CIR.0000000000000644. PMID: 21403094 Free PMC article. Retracted.
-
Cardiac stem cell niches.Stem Cell Res. 2014 Nov;13(3 Pt B):631-46. doi: 10.1016/j.scr.2014.09.001. Epub 2014 Sep 8. Stem Cell Res. 2014. PMID: 25267073 Free PMC article. Review.
-
Improved Generation of Induced Cardiomyocytes Using a Polycistronic Construct Expressing Optimal Ratio of Gata4, Mef2c and Tbx5.J Vis Exp. 2015 Nov 13;(105):53426. doi: 10.3791/53426. J Vis Exp. 2015. PMID: 26649751 Free PMC article.
References
-
- Barker N, et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449:1003–1007. - PubMed
-
- Schmidt M, et al. Clonality analysis after retroviral-mediated gene transfer to CD34+ cells from the cord blood of ADA-deficient SCID neonates. Nat Med. 2003;9:463–468. - PubMed
-
- Mazurier F, Gan OI, McKenzie JL, Doedens M, Dick JE. Lentivector-mediated clonal tracking reveals intrinsic heterogeneity in the human hematopoietic stem cell compartment and culture-induced stem cell impairment. Blood. 2004;103:545–552. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

