Structural insight into the interaction of proteins containing NPF, DPF, and GPF motifs with the C-terminal EH-domain of EHD1
- PMID: 19798736
- PMCID: PMC2821266
- DOI: 10.1002/pro.258
Structural insight into the interaction of proteins containing NPF, DPF, and GPF motifs with the C-terminal EH-domain of EHD1
Abstract
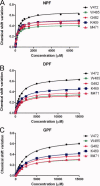
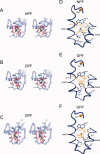
Eps15 homology (EH)-domain containing proteins are regulators of endocytic membrane trafficking. EH-domain binding to proteins containing the tripeptide NPF has been well characterized, but recent studies have shown that EH-domains are also able to interact with ligands containing DPF or GPF motifs. We demonstrate that the three motifs interact in a similar way with the EH-domain of EHD1, with the NPF motif having the highest affinity due to the presence of an intermolecular hydrogen bond. The weaker affinity for the DPF and GPF motifs suggests that if complex formation occurs in vivo, they may require high ligand concentrations, the presence of successive motifs and/or specific flanking residues.
Figures
Similar articles
-
Mechanism for the selective interaction of C-terminal Eps15 homology domain proteins with specific Asn-Pro-Phe-containing partners.J Biol Chem. 2010 Mar 19;285(12):8687-94. doi: 10.1074/jbc.M109.045666. Epub 2010 Jan 27. J Biol Chem. 2010. PMID: 20106972 Free PMC article.
-
Molecular dynamics simulation of the interactions between EHD1 EH domain and multiple peptides.J Zhejiang Univ Sci B. 2015 Oct;16(10):883-96. doi: 10.1631/jzus.B1500106. J Zhejiang Univ Sci B. 2015. PMID: 26465136 Free PMC article.
-
EHD1 and Eps15 interact with phosphatidylinositols via their Eps15 homology domains.J Biol Chem. 2007 Jun 1;282(22):16612-22. doi: 10.1074/jbc.M609493200. Epub 2007 Apr 5. J Biol Chem. 2007. PMID: 17412695
-
Recycling and EH domain proteins at the synapse.Brain Res Brain Res Rev. 2005 Sep;49(2):416-28. doi: 10.1016/j.brainresrev.2005.06.002. Brain Res Brain Res Rev. 2005. PMID: 16054223 Review.
-
Mechanisms of EHD/RME-1 protein function in endocytic transport.Traffic. 2008 Dec;9(12):2043-52. doi: 10.1111/j.1600-0854.2008.00834.x. Epub 2008 Oct 14. Traffic. 2008. PMID: 18801062 Free PMC article. Review.
Cited by
-
Distinct EH domains of the endocytic TPLATE complex confer lipid and protein binding.Nat Commun. 2021 May 24;12(1):3050. doi: 10.1038/s41467-021-23314-6. Nat Commun. 2021. PMID: 34031427 Free PMC article.
-
Differential requirements for the Eps15 homology domain proteins EHD4 and EHD2 in the regulation of mammalian ciliogenesis.Traffic. 2022 Jul;23(7):360-373. doi: 10.1111/tra.12845. Epub 2022 May 17. Traffic. 2022. PMID: 35510564 Free PMC article.
-
Role of the EHD2 unstructured loop in dimerization, protein binding and subcellular localization.PLoS One. 2015 Apr 15;10(4):e0123710. doi: 10.1371/journal.pone.0123710. eCollection 2015. PLoS One. 2015. PMID: 25875965 Free PMC article.
-
Targeting SMYD3 to Sensitize Homologous Recombination-Proficient Tumors to PARP-Mediated Synthetic Lethality.iScience. 2020 Oct 7;23(10):101604. doi: 10.1016/j.isci.2020.101604. eCollection 2020 Oct 23. iScience. 2020. PMID: 33205017 Free PMC article.
-
The enigmatic endosome - sorting the ins and outs of endocytic trafficking.J Cell Sci. 2018 Jul 6;131(13):jcs216499. doi: 10.1242/jcs.216499. J Cell Sci. 2018. PMID: 29980602 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

