The Cdc42 effectors Ste20, Cla4, and Skm1 down-regulate the expression of genes involved in sterol uptake by a mitogen-activated protein kinase-independent pathway
- PMID: 19793923
- PMCID: PMC2777111
- DOI: 10.1091/mbc.e09-01-0034
The Cdc42 effectors Ste20, Cla4, and Skm1 down-regulate the expression of genes involved in sterol uptake by a mitogen-activated protein kinase-independent pathway
Abstract
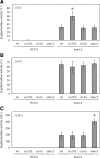
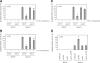
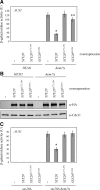
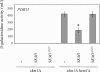
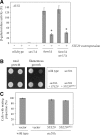
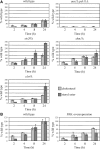
In Saccharomyces cerevisiae, the Rho-type GTPase Cdc42 regulates polarized growth through its effectors, including the p21-activated kinases (PAKs) Ste20, Cla4, and Skm1. Previously, we demonstrated that Ste20 interacts with several proteins involved in sterol synthesis that are crucial for cell polarization. Under anaerobic conditions, sterols cannot be synthesized and need to be imported into cells. Here, we show that Ste20, Cla4, and Skm1 form a complex with Sut1, a transcriptional regulator that promotes sterol uptake. All three PAKs can translocate into the nucleus and down-regulate the expression of genes involved in sterol uptake, including the Sut1 targets AUS1 and DAN1 by a novel mechanism. Consistently, deletion of either STE20, CLA4, or SKM1 results in an increased sterol influx and PAK overexpression inhibits sterol uptake. For Ste20, we demonstrate that the down-regulation of gene expression requires nuclear localization and kinase activity of Ste20. Furthermore, the Ste20-mediated control of expression of sterol uptake genes depends on SUT1 but is independent of a mitogen-activated protein kinase signaling cascade. Together, these observations suggest that PAKs translocate into the nucleus, where they modulate expression of sterol uptake genes via Sut1, thereby controlling sterol homeostasis.
Figures

Similar articles
-
Role of Cdc42-Cla4 interaction in the pheromone response of Saccharomyces cerevisiae.Eukaryot Cell. 2007 Feb;6(2):317-27. doi: 10.1128/EC.00102-06. Epub 2006 Dec 22. Eukaryot Cell. 2007. PMID: 17189484 Free PMC article.
-
Modulation of sterol homeostasis by the Cdc42p effectors Cla4p and Ste20p in the yeast Saccharomyces cerevisiae.FEBS J. 2009 Dec;276(24):7253-64. doi: 10.1111/j.1742-4658.2009.07433.x. FEBS J. 2009. PMID: 20050180
-
Interaction with the SH3 domain protein Bem1 regulates signaling by the Saccharomyces cerevisiae p21-activated kinase Ste20.Mol Cell Biol. 2005 Mar;25(6):2177-90. doi: 10.1128/MCB.25.6.2177-2190.2005. Mol Cell Biol. 2005. PMID: 15743816 Free PMC article.
-
Ste20-related kinases: effectors of signaling and morphogenesis in fungi.Trends Microbiol. 2011 Aug;19(8):400-10. doi: 10.1016/j.tim.2011.04.006. Trends Microbiol. 2011. PMID: 21640592 Review.
-
Regulation of phosphorylation pathways by p21 GTPases. The p21 Ras-related Rho subfamily and its role in phosphorylation signalling pathways.Eur J Biochem. 1996 Dec 1;242(2):171-85. doi: 10.1111/j.1432-1033.1996.0171r.x. Eur J Biochem. 1996. PMID: 8973630 Review.
Cited by
-
A Protein-Protein Interaction Analysis Suggests a Wide Range of New Functions for the p21-Activated Kinase (PAK) Ste20.Int J Mol Sci. 2023 Nov 2;24(21):15916. doi: 10.3390/ijms242115916. Int J Mol Sci. 2023. PMID: 37958899 Free PMC article.
-
The yeast acyltransferase Sct1p regulates fatty acid desaturation by competing with the desaturase Ole1p.Mol Biol Cell. 2012 Apr;23(7):1146-56. doi: 10.1091/mbc.E11-07-0624. Epub 2012 Feb 9. Mol Biol Cell. 2012. PMID: 22323296 Free PMC article.
-
Sulfur Modifications of the Wobble U34 in tRNAs and their Intracellular Localization in Eukaryotic Cells.Biomolecules. 2017 Feb 18;7(1):17. doi: 10.3390/biom7010017. Biomolecules. 2017. PMID: 28218716 Free PMC article. Review.
-
Cell polarization and cytokinesis in budding yeast.Genetics. 2012 Jun;191(2):347-87. doi: 10.1534/genetics.111.132886. Genetics. 2012. PMID: 22701052 Free PMC article. Review.
-
Regulation of vacuolar H+-ATPase activity by the Cdc42 effector Ste20 in Saccharomyces cerevisiae.Eukaryot Cell. 2012 Apr;11(4):442-51. doi: 10.1128/EC.05286-11. Epub 2012 Feb 10. Eukaryot Cell. 2012. PMID: 22327006 Free PMC article.
References
-
- Ahn S. H., Cheung W. L., Hsu J. Y., Diaz R. L., Smith M. M., Allis C. D. Sterile 20 kinase phosphorylates histone H2B at serine 10 during hydrogen peroxide-induced apoptosis in S. cerevisiae. Cell. 2005;120:25–36. - PubMed
-
- Bourot S., Karst F. Isolation and characterization of the Saccharomyces cerevisiae SUT1 gene involved in sterol uptake. Gene. 1995;165:97–102. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

