Structure-function analysis of human immunodeficiency virus type 1 gp120 amino acid mutations associated with resistance to the CCR5 coreceptor antagonist vicriviroc
- PMID: 19776131
- PMCID: PMC2786753
- DOI: 10.1128/JVI.01351-09
Structure-function analysis of human immunodeficiency virus type 1 gp120 amino acid mutations associated with resistance to the CCR5 coreceptor antagonist vicriviroc
Abstract
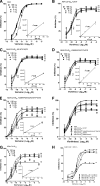
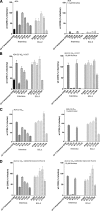
Vicriviroc (VCV) is a small-molecule CCR5 coreceptor antagonist currently in clinical trials for treatment of R5-tropic human immunodeficiency virus type 1 (HIV-1) infection. With this drug in development, identification of resistance mechanisms to VCV is needed to allow optimal outcomes in clinical practice. In this study we further characterized VCV resistance in a lab-adapted, VCV-resistant RU570 virus (RU570-VCV(res)). We show that K305R, R315Q, and K319T amino acid changes in the V3 loop, along with P437S in C4, completely reproduced the resistance phenotype in a chimeric ADA envelope containing the C2-V5 region from RU570 passage control gp120. The K305R amino acid change primarily impacted the degree of resistance, whereas K319T contributed to both resistance and virus infectivity. The P437S mutation in C4 had more influence on the relative degree of virus infectivity, while the R315Q mutation contributed to the virus concentration-dependent phenotypic resistance pattern observed for RU570-VCV(res). RU570-VCV(res) pseudovirus entry with VCV-bound CCR5 was dramatically reduced by Y10A, D11A, Y14A, and Y15A mutations in the N terminus of CCR5, whereas these mutations had less impact on entry in the absence of VCV. Notably, an additional Q315E/I317F substitution in the crown region of the V3 loop enhanced resistance to VCV, resulting in a stronger dependence on the N terminus for viral entry. By fitting the envelope mutations to a molecular model of a recently described docked N-terminal CCR5 peptide consisting of residues 2 to 15 in complex with HIV-1 gp120 CD4, potential new interactions in gp120 with the N terminus of CCR5 were uncovered. The cumulative results of this study suggest that as the RU570 VCV-resistant virus adapted to use the drug-bound receptor, it also developed an increased reliance on the N terminus of CCR5.
Figures

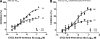

Similar articles
-
Mapping resistance to the CCR5 co-receptor antagonist vicriviroc using heterologous chimeric HIV-1 envelope genes reveals key determinants in the C2-V5 domain of gp120.Virology. 2008 Apr 10;373(2):387-99. doi: 10.1016/j.virol.2007.12.009. Epub 2008 Jan 10. Virology. 2008. PMID: 18190945
-
Clinical resistance to vicriviroc through adaptive V3 loop mutations in HIV-1 subtype D gp120 that alter interactions with the N-terminus and ECL2 of CCR5.Virology. 2010 Apr 25;400(1):145-55. doi: 10.1016/j.virol.2010.01.037. Epub 2010 Feb 21. Virology. 2010. PMID: 20172579
-
HIV-1 clinical isolates resistant to CCR5 antagonists exhibit delayed entry kinetics that are corrected in the presence of drug.J Virol. 2012 Jan;86(2):1119-28. doi: 10.1128/JVI.06421-11. Epub 2011 Nov 16. J Virol. 2012. PMID: 22090117 Free PMC article. Clinical Trial.
-
A pièce de resistance: how HIV-1 escapes small molecule CCR5 inhibitors.Curr Opin HIV AIDS. 2009 Mar;4(2):118-24. doi: 10.1097/COH.0b013e3283223d46. Curr Opin HIV AIDS. 2009. PMID: 19339950 Free PMC article. Review.
-
Genotypic coreceptor analysis.Eur J Med Res. 2007 Oct 15;12(9):453-62. Eur J Med Res. 2007. PMID: 17933727 Review.
Cited by
-
A common mechanism of clinical HIV-1 resistance to the CCR5 antagonist maraviroc despite divergent resistance levels and lack of common gp120 resistance mutations.Retrovirology. 2013 Apr 20;10:43. doi: 10.1186/1742-4690-10-43. Retrovirology. 2013. PMID: 23602046 Free PMC article.
-
Evolution of CCR5 antagonist resistance in an HIV-1 subtype C clinical isolate.J Acquir Immune Defic Syndr. 2010 Dec;55(4):420-7. doi: 10.1097/QAI.0b013e3181f25574. J Acquir Immune Defic Syndr. 2010. PMID: 20856130 Free PMC article.
-
Escape from human immunodeficiency virus type 1 (HIV-1) entry inhibitors.Viruses. 2012 Dec;4(12):3859-911. doi: 10.3390/v4123859. Viruses. 2012. PMID: 23342377 Free PMC article. Review.
-
A maraviroc-resistant HIV-1 with narrow cross-resistance to other CCR5 antagonists depends on both N-terminal and extracellular loop domains of drug-bound CCR5.J Virol. 2010 Oct;84(20):10863-76. doi: 10.1128/JVI.01109-10. Epub 2010 Aug 11. J Virol. 2010. PMID: 20702642 Free PMC article.
-
Distinct HIV-1 entry phenotypes are associated with transmission, subtype specificity, and resistance to broadly neutralizing antibodies.Retrovirology. 2014 Jun 23;11:48. doi: 10.1186/1742-4690-11-48. Retrovirology. 2014. PMID: 24957778 Free PMC article.
References
-
- Blanpain, C., B. Lee, J. Vakili, B. J. Doranz, C. Govaerts, I. Migeotte, M. Sharron, V. Dupriez, G. Vassart, R. W. Doms, and M. Parmentier. 1999. Extracellular cysteines of CCR5 are required for chemokine binding, but dispensable for HIV-1 coreceptor activity. J. Biol. Chem. 274:18902-18908. - PubMed
-
- Chan, D. C., D. Fass, J. M. Berger, and P. S. Kim. 1997. Core structure of gp41 from the HIV envelope glycoprotein. Cell 89:263-273. - PubMed
-
- Chan, D. C., and P. S. Kim. 1998. HIV entry and its inhibition. Cell 93:681-684. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

