Genome-wide expression profiling of in vivo-derived bloodstream parasite stages and dynamic analysis of mRNA alterations during synchronous differentiation in Trypanosoma brucei
- PMID: 19747379
- PMCID: PMC2753553
- DOI: 10.1186/1471-2164-10-427
Genome-wide expression profiling of in vivo-derived bloodstream parasite stages and dynamic analysis of mRNA alterations during synchronous differentiation in Trypanosoma brucei
Abstract
Background: Trypanosomes undergo extensive developmental changes during their complex life cycle. Crucial among these is the transition between slender and stumpy bloodstream forms and, thereafter, the differentiation from stumpy to tsetse-midgut procyclic forms. These developmental events are highly regulated, temporally reproducible and accompanied by expression changes mediated almost exclusively at the post-transcriptional level.
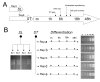
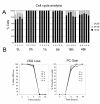
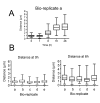
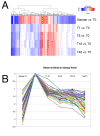
Results: In this study we have examined, by whole-genome microarray analysis, the mRNA abundance of genes in slender and stumpy forms of T.brucei AnTat1.1 cells, and also during their synchronous differentiation to procyclic forms. In total, five biological replicates representing the differentiation of matched parasite populations derived from five individual mouse infections were assayed, with RNAs being derived at key biological time points during the time course of their synchronous differentiation to procyclic forms. Importantly, the biological context of these mRNA profiles was established by assaying the coincident cellular events in each population (surface antigen exchange, morphological restructuring, cell cycle re-entry), thereby linking the observed gene expression changes to the well-established framework of trypanosome differentiation.
Conclusion: Using stringent statistical analysis and validation of the derived profiles against experimentally-predicted gene expression and phenotypic changes, we have established the profile of regulated gene expression during these important life-cycle transitions. The highly synchronous nature of differentiation between stumpy and procyclic forms also means that these studies of mRNA profiles are directly relevant to the changes in mRNA abundance within individual cells during this well-characterised developmental transition.
Figures

Similar articles
-
Regulation of Trypanosoma brucei Total and Polysomal mRNA during Development within Its Mammalian Host.PLoS One. 2013 Jun 26;8(6):e67069. doi: 10.1371/journal.pone.0067069. Print 2013. PLoS One. 2013. PMID: 23840587 Free PMC article.
-
RNA-Seq analysis validates the use of culture-derived Trypanosoma brucei and provides new markers for mammalian and insect life-cycle stages.BMC Genomics. 2018 Apr 2;19(1):227. doi: 10.1186/s12864-018-4600-6. BMC Genomics. 2018. PMID: 29606092 Free PMC article.
-
Extensive stage-regulation of translation revealed by ribosome profiling of Trypanosoma brucei.BMC Genomics. 2014 Oct 20;15(1):911. doi: 10.1186/1471-2164-15-911. BMC Genomics. 2014. PMID: 25331479 Free PMC article.
-
Bloodstream form pre-adaptation to the tsetse fly in Trypanosoma brucei.Front Cell Infect Microbiol. 2013 Nov 14;3:78. doi: 10.3389/fcimb.2013.00078. eCollection 2013. Front Cell Infect Microbiol. 2013. PMID: 24294594 Free PMC article. Review.
-
Life-cycle differentiation in Trypanosoma brucei: molecules and mutants.Biochem Soc Trans. 2000 Oct;28(5):531-6. doi: 10.1042/bst0280531. Biochem Soc Trans. 2000. PMID: 11044369 Review.
Cited by
-
P-type transport ATPases in Leishmania and Trypanosoma.Parasite. 2019;26:69. doi: 10.1051/parasite/2019069. Epub 2019 Nov 29. Parasite. 2019. PMID: 31782726 Free PMC article. Review.
-
Molecular control of irreversible bistability during trypanosome developmental commitment.J Cell Biol. 2015 Oct 26;211(2):455-68. doi: 10.1083/jcb.201506114. Epub 2015 Oct 19. J Cell Biol. 2015. PMID: 26483558 Free PMC article.
-
Trypanosoma brucei metabolism is under circadian control.Nat Microbiol. 2017 Mar 13;2:17032. doi: 10.1038/nmicrobiol.2017.32. Nat Microbiol. 2017. PMID: 28288095 Free PMC article.
-
Lipidomic analysis of bloodstream and procyclic form Trypanosoma brucei.Parasitology. 2010 Aug;137(9):1357-92. doi: 10.1017/S0031182010000715. Parasitology. 2010. PMID: 20602846 Free PMC article. Review.
-
Transcriptome analysis of differentiating trypanosomes reveals the existence of multiple post-transcriptional regulons.BMC Genomics. 2009 Oct 26;10:495. doi: 10.1186/1471-2164-10-495. BMC Genomics. 2009. PMID: 19857263 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

