A genome-wide genetic screen for host factors required for hepatitis C virus propagation
- PMID: 19717417
- PMCID: PMC2752535
- DOI: 10.1073/pnas.0907439106
A genome-wide genetic screen for host factors required for hepatitis C virus propagation
Abstract
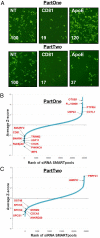
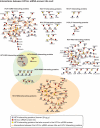
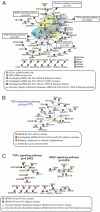
Hepatitis C virus (HCV) infection is a major cause of end-stage liver disease and a leading indication for liver transplantation. Current therapy fails in many instances and is associated with significant side effects. HCV encodes only a few proteins and depends heavily on host factors for propagation. Each of these host dependencies is a potential therapeutic target. To find host factors required by HCV, we completed a genome-wide small interfering RNA (siRNA) screen using an infectious HCV cell culture system. We applied a two-part screening protocol to allow identification of host factors involved in the complete viral lifecycle. The candidate genes found included known or previously identified factors, and also implicate many additional host cell proteins in HCV infection. To create a more comprehensive view of HCV and host cell interactions, we performed a bioinformatic meta-analysis that integrates our data with those of previous functional and proteomic studies. The identification of host factors participating in the complete HCV lifecycle will both advance our understanding of HCV pathogenesis and illuminate therapeutic targets.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Integrative functional genomics of hepatitis C virus infection identifies host dependencies in complete viral replication cycle.PLoS Pathog. 2014 May 22;10(5):e1004163. doi: 10.1371/journal.ppat.1004163. eCollection 2014 May. PLoS Pathog. 2014. PMID: 24852294 Free PMC article.
-
Identification and comparative analysis of hepatitis C virus-host cell protein interactions.Mol Biosyst. 2013 Dec;9(12):3199-209. doi: 10.1039/c3mb70343f. Epub 2013 Oct 18. Mol Biosyst. 2013. PMID: 24136289 Free PMC article.
-
Involvement of creatine kinase B in hepatitis C virus genome replication through interaction with the viral NS4A protein.J Virol. 2009 May;83(10):5137-47. doi: 10.1128/JVI.02179-08. Epub 2009 Mar 4. J Virol. 2009. PMID: 19264780 Free PMC article.
-
Mechanisms involved in the development of chronic hepatitis C as potential targets of antiviral therapy.Curr Pharm Biotechnol. 2011 Nov;12(11):1774-80. doi: 10.2174/138920111798377030. Curr Pharm Biotechnol. 2011. PMID: 21902631 Review.
-
High-throughput approaches to unravel hepatitis C virus-host interactions.Virus Res. 2016 Jun 15;218:18-24. doi: 10.1016/j.virusres.2015.09.013. Epub 2015 Sep 26. Virus Res. 2016. PMID: 26410623 Review.
Cited by
-
RNA-sequencing analysis of 5' capped RNAs identifies many new differentially expressed genes in acute hepatitis C virus infection.Viruses. 2012 Apr;4(4):581-612. doi: 10.3390/v4040581. Epub 2012 Apr 16. Viruses. 2012. PMID: 22590687 Free PMC article.
-
Lipids and HCV.Semin Immunopathol. 2013 Jan;35(1):87-100. doi: 10.1007/s00281-012-0356-2. Epub 2012 Oct 31. Semin Immunopathol. 2013. PMID: 23111699 Review.
-
The Power Decoder Simulator for the Evaluation of Pooled shRNA Screen Performance.J Biomol Screen. 2015 Sep;20(8):965-75. doi: 10.1177/1087057115576715. Epub 2015 Mar 16. J Biomol Screen. 2015. PMID: 25777298 Free PMC article.
-
Tyrphostin AG1478 Inhibits Encephalomyocarditis Virus and Hepatitis C Virus by Targeting Phosphatidylinositol 4-Kinase IIIα.Antimicrob Agents Chemother. 2016 Sep 23;60(10):6402-6. doi: 10.1128/AAC.01331-16. Print 2016 Oct. Antimicrob Agents Chemother. 2016. PMID: 27480860 Free PMC article.
-
The dependence of viral RNA replication on co-opted host factors.Nat Rev Microbiol. 2011 Dec 19;10(2):137-49. doi: 10.1038/nrmicro2692. Nat Rev Microbiol. 2011. PMID: 22183253 Free PMC article. Review.
References
-
- Moradpour D, Penin F, Rice CM. Replication of hepatitis C virus. Nat Rev Microbiol. 2007;5:453–463. - PubMed
-
- Lindenbach BD, Rice CM. Unravelling hepatitis C virus replication from genome to function. Nature. 2005;436:933–938. - PubMed
-
- Miyanari Y, et al. The lipid droplet is an important organelle for hepatitis C virus production. Nat Cell Biol. 2007;9:1089–1097. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

