Lin28 recruits the TUTase Zcchc11 to inhibit let-7 maturation in mouse embryonic stem cells
- PMID: 19713958
- PMCID: PMC2758923
- DOI: 10.1038/nsmb.1676
Lin28 recruits the TUTase Zcchc11 to inhibit let-7 maturation in mouse embryonic stem cells
Abstract
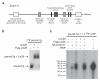
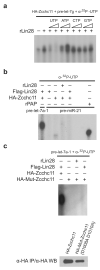
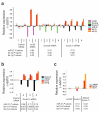
Lin28 and Lin28B, two developmentally regulated RNA-binding proteins and likely proto-oncogenes, selectively inhibit the maturation of let-7 family microRNAs (miRNAs) in embryonic stem cells and certain cancer cell lines. Moreover, let-7 precursors (pre-let-7) were previously found to be terminally uridylated in a Lin28-dependent fashion. Here we identify Zcchc11 (zinc finger, CCHC domain containing 11) as the 3' terminal uridylyl transferase (TUTase) responsible for Lin28-mediated pre-let-7 uridylation and subsequent blockade of let-7 processing in mouse embryonic stem cells. We demonstrate that Zcchc11 activity is UTP-dependent, selective for let-7 and recruited by Lin28. Furthermore, knockdown of either Zcchc11 or Lin28, or overexpression of a catalytically inactive TUTase, relieves the selective inhibition of let-7 processing and leads to the accumulation of mature let-7 miRNAs and repression of let-7 target reporter genes. Our results establish a role for Zcchc11-catalyzed pre-let-7 uridylation in the control of miRNA biogenesis.
Figures
Similar articles
-
Identification of small molecule inhibitors of Zcchc11 TUTase activity.RNA Biol. 2015;12(8):792-800. doi: 10.1080/15476286.2015.1058478. RNA Biol. 2015. PMID: 26114892 Free PMC article.
-
Lin28-mediated control of let-7 microRNA expression by alternative TUTases Zcchc11 (TUT4) and Zcchc6 (TUT7).RNA. 2012 Oct;18(10):1875-85. doi: 10.1261/rna.034538.112. Epub 2012 Aug 16. RNA. 2012. PMID: 22898984 Free PMC article.
-
Lin28 mediates the terminal uridylation of let-7 precursor MicroRNA.Mol Cell. 2008 Oct 24;32(2):276-84. doi: 10.1016/j.molcel.2008.09.014. Mol Cell. 2008. PMID: 18951094
-
The LIN28/let-7 Pathway in Cancer.Front Genet. 2017 Mar 28;8:31. doi: 10.3389/fgene.2017.00031. eCollection 2017. Front Genet. 2017. PMID: 28400788 Free PMC article. Review.
-
A mirror of two faces: Lin28 as a master regulator of both miRNA and mRNA.Wiley Interdiscip Rev RNA. 2012 Jul-Aug;3(4):483-94. doi: 10.1002/wrna.1112. Epub 2012 Mar 29. Wiley Interdiscip Rev RNA. 2012. PMID: 22467269 Review.
Cited by
-
microRNAs associated with the different human Argonaute proteins.Nucleic Acids Res. 2012 Oct;40(19):9850-62. doi: 10.1093/nar/gks705. Epub 2012 Jul 25. Nucleic Acids Res. 2012. PMID: 22844086 Free PMC article.
-
Improved crystallization and diffraction of caffeine-induced death suppressor protein 1 (Cid1).Acta Crystallogr F Struct Biol Commun. 2015 Mar;71(Pt 3):346-53. doi: 10.1107/S2053230X15001351. Epub 2015 Feb 21. Acta Crystallogr F Struct Biol Commun. 2015. PMID: 25760713 Free PMC article.
-
Uridylation of RNA Hairpins by Tailor Confines the Emergence of MicroRNAs in Drosophila.Mol Cell. 2015 Jul 16;59(2):203-16. doi: 10.1016/j.molcel.2015.05.033. Epub 2015 Jul 2. Mol Cell. 2015. PMID: 26145176 Free PMC article.
-
Responsive fluorescent nucleotides serve as efficient substrates to probe terminal uridylyl transferase.Chem Commun (Camb). 2020 Oct 21;56(82):12319-12322. doi: 10.1039/d0cc05092j. Epub 2020 Sep 17. Chem Commun (Camb). 2020. PMID: 32939524 Free PMC article.
-
MicroRNA biogenesis: regulating the regulators.Crit Rev Biochem Mol Biol. 2013 Jan-Feb;48(1):51-68. doi: 10.3109/10409238.2012.738643. Epub 2012 Nov 19. Crit Rev Biochem Mol Biol. 2013. PMID: 23163351 Free PMC article. Review.
References
-
- Winter J, Jung S, Keller S, Gregory RI, Diederichs S. Many roads to maturity: microRNA biogenesis pathways and their regulation. Nat Cell Biol. 2009;11:228–34. - PubMed
-
- Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–69. - PubMed
-
- Hagan JP, Croce CM. MicroRNAs in carcinogenesis. Cytogenet Genome Res. 2007;118:252–9. - PubMed
-
- Bussing I, Slack FJ, Grosshans H. let-7 microRNAs in development, stem cells and cancer. Trends Mol Med. 2008;14:400–9. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

