The highly attenuated oncolytic recombinant vaccinia virus GLV-1h68: comparative genomic features and the contribution of F14.5L inactivation
- PMID: 19701652
- PMCID: PMC2746888
- DOI: 10.1007/s00438-009-0475-1
The highly attenuated oncolytic recombinant vaccinia virus GLV-1h68: comparative genomic features and the contribution of F14.5L inactivation
Abstract
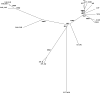
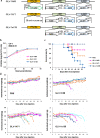
As a new anticancer treatment option, vaccinia virus (VACV) has shown remarkable antitumor activities (oncolysis) in preclinical studies, but potential infection of other organs remains a safety concern. We present here genome comparisons between the de novo sequence of GLV-1h68, a recombinant VACV, and other VACVs. The identified differences in open reading frames (ORFs) include genes encoding host-range selection, virulence and immune modulation proteins, e.g., ankyrin-like proteins, serine proteinase inhibitor SPI-2/CrmA, tumor necrosis factor (TNF) receptor homolog CrmC, semaphorin-like and interleukin-1 receptor homolog proteins. Phylogenetic analyses indicate that GLV-1h68 is closest to Lister strains but has lost several ORFs present in its parental LIVP strain, including genes encoding CrmE and a viral Golgi anti-apoptotic protein, v-GAAP. The reduced pathogenicity of GLV-1h68 is confirmed in male mice bearing C6 rat glioma and in immunocompetent mice bearing B16-F10 murine melanoma. The contribution of foreign gene expression cassettes in the F14.5L, J2R and A56R loci is analyzed, in particular the contribution of F14.5L inactivation to the reduced virulence is demonstrated by comparing the virulence of GLV-1h68 with its F14.5L-null and revertant viruses. GLV-1h68 is a promising engineered VACV variant for anticancer therapy with tumor-specific replication, reduced pathogenicity and benign tissue tropism.
Figures





Similar articles
-
Replication efficiency of oncolytic vaccinia virus in cell cultures prognosticates the virulence and antitumor efficacy in mice.J Transl Med. 2011 Sep 27;9:164. doi: 10.1186/1479-5876-9-164. J Transl Med. 2011. PMID: 21951588 Free PMC article.
-
Growth inhibition of different human colorectal cancer xenografts after a single intravenous injection of oncolytic vaccinia virus GLV-1h68.J Transl Med. 2013 Mar 26;11:79. doi: 10.1186/1479-5876-11-79. J Transl Med. 2013. PMID: 23531320 Free PMC article.
-
Eradication of solid human breast tumors in nude mice with an intravenously injected light-emitting oncolytic vaccinia virus.Cancer Res. 2007 Oct 15;67(20):10038-46. doi: 10.1158/0008-5472.CAN-07-0146. Cancer Res. 2007. PMID: 17942938
-
Efficient colonization and therapy of human hepatocellular carcinoma (HCC) using the oncolytic vaccinia virus strain GLV-1h68.PLoS One. 2011;6(7):e22069. doi: 10.1371/journal.pone.0022069. Epub 2011 Jul 11. PLoS One. 2011. PMID: 21779374 Free PMC article.
-
The complete genomic sequence of the modified vaccinia Ankara strain: comparison with other orthopoxviruses.Virology. 1998 May 10;244(2):365-96. doi: 10.1006/viro.1998.9123. Virology. 1998. PMID: 9601507 Review.
Cited by
-
Oncolytic vaccinia virus GLV-1h68 exhibits profound antitumoral activities in cell lines originating from neuroendocrine neoplasms.BMC Cancer. 2020 Jul 6;20(1):628. doi: 10.1186/s12885-020-07121-8. BMC Cancer. 2020. PMID: 32631270 Free PMC article.
-
Comprehensive assessment on the applications of oncolytic viruses for cancer immunotherapy.Front Pharmacol. 2022 Dec 8;13:1082797. doi: 10.3389/fphar.2022.1082797. eCollection 2022. Front Pharmacol. 2022. PMID: 36569326 Free PMC article. Review.
-
Genetic ancestry and population structure of vaccinia virus.NPJ Vaccines. 2022 Aug 11;7(1):92. doi: 10.1038/s41541-022-00519-4. NPJ Vaccines. 2022. PMID: 35953491 Free PMC article.
-
Evolution of and evolutionary relationships between extant vaccinia virus strains.J Virol. 2015 Feb;89(3):1809-24. doi: 10.1128/JVI.02797-14. Epub 2014 Nov 19. J Virol. 2015. PMID: 25410873 Free PMC article.
-
Current status of gene therapy for brain tumors.Transl Res. 2013 Apr;161(4):339-54. doi: 10.1016/j.trsl.2012.11.003. Epub 2012 Dec 11. Transl Res. 2013. PMID: 23246627 Free PMC article.
References
-
- {'text': '', 'ref_index': 1, 'ids': [{'type': 'PubMed', 'value': '4092585', 'is_inner': True, 'url': 'https://pubmed.ncbi.nlm.nih.gov/4092585/'}]}
- Al’tshtein AD, Zakharova LG, Loparev VN, Pashvykina GV, Gorodetskii SI (1985) Isolation of a recombinant vaccinia virus based on the LIVP strain inducing the surface antigen of the hepatitis B virus. Dokl Akad Nauk SSSR 285:696–699 - PubMed
-
- {'text': '', 'ref_index': 1, 'ids': [{'type': 'PubMed', 'value': '10211965', 'is_inner': True, 'url': 'https://pubmed.ncbi.nlm.nih.gov/10211965/'}]}
- Alcami A, Khanna A, Paul NL, Smith GL (1999) Vaccinia virus strains Lister, USSR and Evans express soluble and cell-surface tumour necrosis factor receptors. J Gen Virol 80:949–959 - PubMed
-
- {'text': '', 'ref_index': 1, 'ids': [{'type': 'DOI', 'value': '10.1006/viro.1998.9123', 'is_inner': False, 'url': 'https://doi.org/10.1006/viro.1998.9123'}, {'type': 'PubMed', 'value': '9601507', 'is_inner': True, 'url': 'https://pubmed.ncbi.nlm.nih.gov/9601507/'}]}
- Antoine G, Scheiflinger F, Dorner F, Falkner FG (1998) The complete genomic sequence of the modified vaccinia Ankara strain: comparison with other orthopoxviruses. Virology 244:365–396 - PubMed
-
- {'text': '', 'ref_index': 1, 'ids': [{'type': 'DOI', 'value': '10.1093/nar/gkh121', 'is_inner': False, 'url': 'https://doi.org/10.1093/nar/gkh121'}, {'type': 'PMC', 'value': 'PMC308855', 'is_inner': False, 'url': 'https://pmc.ncbi.nlm.nih.gov/articles/PMC308855/'}, {'type': 'PubMed', 'value': '14681378', 'is_inner': True, 'url': 'https://pubmed.ncbi.nlm.nih.gov/14681378/'}]}
- Bateman A, Coin L, Durbin R, Finn RD, Hollich V, Griffiths-Jones S, Khanna A, Marshall M, Moxon S et al (2004) The Pfam protein families database. Nucl Acid Res 32:D138–D141 - PMC - PubMed
-
- {'text': '', 'ref_index': 1, 'ids': [{'type': 'DOI', 'value': '10.1093/nar/gki487', 'is_inner': False, 'url': 'https://doi.org/10.1093/nar/gki487'}, {'type': 'PMC', 'value': 'PMC1160247', 'is_inner': False, 'url': 'https://pmc.ncbi.nlm.nih.gov/articles/PMC1160247/'}, {'type': 'PubMed', 'value': '15980510', 'is_inner': True, 'url': 'https://pubmed.ncbi.nlm.nih.gov/15980510/'}]}
- Besemer J, Borodovsky M (2005) GeneMark: web software for gene finding in prokaryotes, eukaryotes and viruses. Nucl Acid Res 33:W451–W454 - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

