Cytomegalovirus promoter up-regulation is the major cause of increased protein levels of unstable reporter proteins after treatment of living cells with proteasome inhibitors
- PMID: 19679666
- PMCID: PMC2788877
- DOI: 10.1074/jbc.M109.004101
Cytomegalovirus promoter up-regulation is the major cause of increased protein levels of unstable reporter proteins after treatment of living cells with proteasome inhibitors
Retraction in
-
Cytomegalovirus promoter up-regulation is the major cause of increased protein levels of unstable reporter proteins after treatment of living cells with proteasome inhibitors.J Biol Chem. 2016 Apr 22;291(17):8985. doi: 10.1074/jbc.A109.004101. J Biol Chem. 2016. PMID: 27129194 Free PMC article. No abstract available.
Abstract
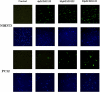
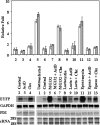
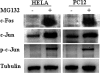
Fluorescent unstable proteins obtained by the fusion of a fluorescent protein coding sequence with specific amino acid sequences that promote its fast degradation have become popular to gauge the activity of the ubiquitin/proteasome system in living cells. The steady-state levels of expression of these unstable proteins is low in agreement with their short half-lives, and they accumulate in the cell upon treatment with proteasome inhibitors. We show here that this accumulation is mainly due to transcriptional up-regulation of the cytomegalovirus promoter by proteasome inhibitors and mediated, at least in part, by AP1 transactivation. These simple facts put under quarantine conclusions reached about the activity of the ubiquitin/proteasome pathway in animal cells in culture or in transgenic mice, where popular cytomegalovirus-driven constructs are used, as transcriptional regulation of the expression of the reporter protein construct and not degradation of the unstable protein by the ubiquitin/proteasome system may contribute significantly to the interpretation of the results observed.
Figures






Similar articles
-
Proteasome inhibitor differentially regulates expression of the major immediate early genes of human cytomegalovirus in human central nervous system-derived cell lines.Virus Res. 2009 Jun;142(1-2):68-77. doi: 10.1016/j.virusres.2009.01.010. Epub 2009 Feb 6. Virus Res. 2009. PMID: 19201384
-
PEST motif sequence regulating human NANOG for proteasomal degradation.Stem Cells Dev. 2011 Sep;20(9):1511-9. doi: 10.1089/scd.2010.0410. Epub 2011 Mar 12. Stem Cells Dev. 2011. PMID: 21299413
-
Conditional transgenic system for mouse aurora a kinase: degradation by the ubiquitin proteasome pathway controls the level of the transgenic protein.Mol Cell Biol. 2005 Jun;25(12):5270-81. doi: 10.1128/MCB.25.12.5270-5281.2005. Mol Cell Biol. 2005. PMID: 15923640 Free PMC article.
-
Proteasome inhibitors and cardiac cell growth.Cardiovasc Res. 2010 Jan 15;85(2):321-9. doi: 10.1093/cvr/cvp226. Epub 2009 Jul 3. Cardiovasc Res. 2010. PMID: 19578073 Free PMC article. Review.
-
The ubiquitin-proteasome system as a drug target in cerebrovascular disease: therapeutic potential of proteasome inhibitors.Curr Opin Investig Drugs. 2005 Jul;6(7):686-99. Curr Opin Investig Drugs. 2005. PMID: 16044664 Free PMC article. Review.
Cited by
-
A photoconvertible reporter of the ubiquitin-proteasome system in vivo.Nat Methods. 2010 Jun;7(6):473-8. doi: 10.1038/nmeth.1460. Epub 2010 May 9. Nat Methods. 2010. PMID: 20453865
-
Indirect inhibition of 26S proteasome activity in a cellular model of Huntington's disease.J Cell Biol. 2012 Mar 5;196(5):573-87. doi: 10.1083/jcb.201110093. Epub 2012 Feb 27. J Cell Biol. 2012. PMID: 22371559 Free PMC article.
-
Human cytomegalovirus IE1 protein disrupts interleukin-6 signaling by sequestering STAT3 in the nucleus.J Virol. 2013 Oct;87(19):10763-76. doi: 10.1128/JVI.01197-13. Epub 2013 Jul 31. J Virol. 2013. PMID: 23903834 Free PMC article.
-
Calreticulin and Arginylated Calreticulin Have Different Susceptibilities to Proteasomal Degradation.J Biol Chem. 2015 Jun 26;290(26):16403-14. doi: 10.1074/jbc.M114.626127. Epub 2015 May 12. J Biol Chem. 2015. PMID: 25969538 Free PMC article.
-
Quantitative cell-based protein degradation assays to identify and classify drugs that target the ubiquitin-proteasome system.J Biol Chem. 2011 May 13;286(19):16546-54. doi: 10.1074/jbc.M110.215319. Epub 2011 Feb 22. J Biol Chem. 2011. PMID: 21343295 Free PMC article.
References
-
- Voges D., Zwickl P., Baumeister W. (1999) Annu. Rev. Biochem. 68, 1015–1068 - PubMed
-
- Hershko A., Ciechanover A. (1998) Annu. Rev. Biochem. 67, 425–479 - PubMed
-
- Varshavsky A. (1995) Cold. Spring. Harb. Symp. Quant. B 60, 461–478 - PubMed
-
- Dantuma N. P., Lindsten K., Glas R., Jellne M., Masucci M. G. (2000) Nat. Biotechnol. 18, 538–543 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

