Essential global role of CDC14 in DNA synthesis revealed by chromosome underreplication unrecognized by checkpoints in cdc14 mutants
- PMID: 19666479
- PMCID: PMC2723162
- DOI: 10.1073/pnas.0900190106
Essential global role of CDC14 in DNA synthesis revealed by chromosome underreplication unrecognized by checkpoints in cdc14 mutants
Abstract
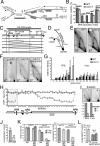
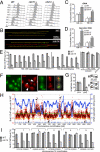
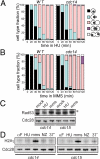
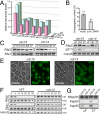
The CDC14 family of multifunctional evolutionarily conserved phosphatases includes major regulators of mitosis in eukaryotes and of DNA damage response in humans. The CDC14 function is also crucial for accurate chromosome segregation, which is exemplified by its absolute requirement in yeast for the anaphase segregation of nucleolar organizers; however the nature of this essential pathway is not understood. Upon investigation of the rDNA nondisjunction phenomenon, it was found that cdc14 mutants fail to complete replication of this locus. Moreover, other late-replicating genomic regions (10% of the genome) are also underreplicated in cdc14 mutants undergoing anaphase. This selective genome-wide replication defect is due to dosage insufficiency of replication factors in the nucleus, which stems from two defects, both contingent on the reduced CDC14 function in the preceding mitosis. First, a constitutive nuclear import defect results in a drastic dosage decrease for those replication proteins that are regulated by nuclear transport. Particularly, essential RPA subunits display both lower mRNA and protein levels, as well as abnormal cytoplasmic localization. Second, the reduced transcription of MBF and SBF-controlled genes in G1 leads to the reduction in protein levels of many proteins involved in DNA replication. The failure to complete replication of late replicons is the primary reason for chromosome nondisjunction upon CDC14 dysfunction. As the genome-wide slow-down of DNA replication does not trigger checkpoints [Lengronne A, Schwob E (2002) Mol Cell 9:1067-1078], CDC14 mutations pose an overwhelming challenge to genome stability, both generating chromosome damage and undermining the checkpoint control mechanisms.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
Anaphase onset before complete DNA replication with intact checkpoint responses.Science. 2007 Mar 9;315(5817):1411-5. doi: 10.1126/science.1134025. Science. 2007. PMID: 17347440
-
Cdc14 phosphatase resolves the rDNA segregation delay.Nat Cell Biol. 2004 Jun;6(6):473-5. doi: 10.1038/ncb0604-473. Nat Cell Biol. 2004. PMID: 15170454
-
Ribosomal DNA replication time coordinates completion of genome replication and anaphase in yeast.Cell Rep. 2023 Mar 28;42(3):112161. doi: 10.1016/j.celrep.2023.112161. Epub 2023 Feb 25. Cell Rep. 2023. PMID: 36842087 Free PMC article.
-
Cdc14 phosphatase: warning, no delay allowed for chromosome segregation!Curr Genet. 2016 Feb;62(1):7-13. doi: 10.1007/s00294-015-0502-1. Epub 2015 Jun 27. Curr Genet. 2016. PMID: 26116076 Free PMC article. Review.
-
Cdc14 and the temporal coordination between mitotic exit and chromosome segregation.Cell Cycle. 2005 Jan;4(1):109-12. doi: 10.4161/cc.4.1.1356. Epub 2005 Jan 10. Cell Cycle. 2005. PMID: 15611663 Review.
Cited by
-
Pif1-family helicases cooperatively suppress widespread replication-fork arrest at tRNA genes.Nat Struct Mol Biol. 2017 Feb;24(2):162-170. doi: 10.1038/nsmb.3342. Epub 2016 Dec 19. Nat Struct Mol Biol. 2017. PMID: 27991904 Free PMC article.
-
DNA replication timing: Biochemical mechanisms and biological significance.Bioessays. 2022 Nov;44(11):e2200097. doi: 10.1002/bies.202200097. Epub 2022 Sep 20. Bioessays. 2022. PMID: 36125226 Free PMC article.
-
The Cdc14 Phosphatase Controls Resolution of Recombination Intermediates and Crossover Formation during Meiosis.Int J Mol Sci. 2021 Sep 10;22(18):9811. doi: 10.3390/ijms22189811. Int J Mol Sci. 2021. PMID: 34575966 Free PMC article.
-
Hst3p, a histone deacetylase, promotes maintenance of Saccharomyces cerevisiae chromosome III lacking efficient replication origins.Mol Genet Genomics. 2016 Feb;291(1):271-83. doi: 10.1007/s00438-015-1105-8. Epub 2015 Aug 29. Mol Genet Genomics. 2016. PMID: 26319649 Free PMC article.
-
Cancer models in Caenorhabditis elegans.Dev Dyn. 2010 May;239(5):1413-48. doi: 10.1002/dvdy.22247. Dev Dyn. 2010. PMID: 20175192 Free PMC article. Review.
References
-
- Stegmeier F, Amon A. Closing mitosis: The functions of the Cdc14 phosphatase and its regulation. Annu Rev Genet. 2004;38:203–232. - PubMed
-
- Trautmann S, et al. The S. pombe Cdc14-like phosphatase Clp1p regulates chromosome biorientation and interacts with Aurora kinase. Dev Cell. 2004;7:755–762. - PubMed
-
- Krasinska L, et al. Regulation of multiple cell cycle events by Cdc14 homologues in vertebrates. Exp Cell Res. 2007;313:1225–1239. - PubMed
-
- Visintin R, et al. Cfi1 prevents premature exit from mitosis by anchoring Cdc14 phosphatase in the nucleolus. Nature. 1999;398:818–823. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

