Open reading frame 33 of a gammaherpesvirus encodes a tegument protein essential for virion morphogenesis and egress
- PMID: 19656880
- PMCID: PMC2753129
- DOI: 10.1128/JVI.00497-09
Open reading frame 33 of a gammaherpesvirus encodes a tegument protein essential for virion morphogenesis and egress
Abstract
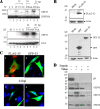
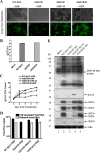
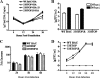
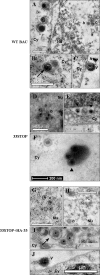
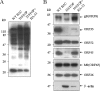
Tegument is a unique structure of herpesvirus, which surrounds the capsid and interacts with the envelope. Morphogenesis of gammaherpesvirus is poorly understood due to lack of efficient lytic replication for Epstein-Barr virus and Kaposi's sarcoma-associated herpesvirus/human herpesvirus 8, which are etiologically associated with several types of human malignancies. Murine gammaherpesvirus 68 (MHV-68) is genetically related to the human gammaherpesviruses and presents an excellent model for studying de novo lytic replication of gammaherpesviruses. MHV-68 open reading frame 33 (ORF33) is conserved among Alpha-, Beta-, and Gammaherpesvirinae subfamilies. However, the specific role of ORF33 in gammaherpesvirus replication has not yet been characterized. We describe here that ORF33 is a true late gene and encodes a tegument protein. By constructing an ORF33-null MHV-68 mutant, we demonstrated that ORF33 is not required for viral DNA replication, early and late gene expression, viral DNA packaging or capsid assembly but is required for virion morphogenesis and egress. Although the ORF33-null virus was deficient in release of infectious virions, partially tegumented capsids produced by the ORF33-null mutant accumulated in the cytoplasm, containing conserved capsid proteins, ORF52 tegument protein, but virtually no ORF45 tegument protein and the 65-kDa glycoprotein B. Finally, we found that the defect of ORF33-null MHV-68 could be rescued by providing ORF33 in trans or in an ORF33-null revertant virus. Taken together, our results indicate that ORF33 is a tegument protein required for viral lytic replication and functions in virion morphogenesis and egress.
Figures

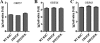
Similar articles
-
Gammaherpesvirus Tegument Protein ORF33 Is Associated With Intranuclear Capsids at an Early Stage of the Tegumentation Process.J Virol. 2015 May;89(10):5288-97. doi: 10.1128/JVI.00079-15. Epub 2015 Feb 25. J Virol. 2015. PMID: 25717105 Free PMC article.
-
The Interaction between Tegument Proteins ORF33 and ORF45 Plays an Essential Role in Cytoplasmic Virion Maturation of a Gammaherpesvirus.J Virol. 2022 Nov 23;96(22):e0107322. doi: 10.1128/jvi.01073-22. Epub 2022 Oct 27. J Virol. 2022. PMID: 36300940 Free PMC article.
-
Murine gammaherpesvirus 68 ORF52 encodes a tegument protein required for virion morphogenesis in the cytoplasm.J Virol. 2007 Sep;81(18):10137-50. doi: 10.1128/JVI.01233-06. Epub 2007 Jul 18. J Virol. 2007. PMID: 17634243 Free PMC article.
-
Comprehensive Analysis of the Tegument Proteins Involved in Capsid Transport and Virion Morphogenesis of Alpha, Beta and Gamma Herpesviruses.Viruses. 2023 Oct 6;15(10):2058. doi: 10.3390/v15102058. Viruses. 2023. PMID: 37896835 Free PMC article. Review.
-
Role of tegument proteins in herpesvirus assembly and egress.Protein Cell. 2010 Nov;1(11):987-98. doi: 10.1007/s13238-010-0120-0. Epub 2010 Dec 10. Protein Cell. 2010. PMID: 21153516 Free PMC article. Review.
Cited by
-
The ORF45 Protein of Kaposi's Sarcoma-Associated Herpesvirus and Its Critical Role in the Viral Life Cycle.Viruses. 2022 Sep 11;14(9):2010. doi: 10.3390/v14092010. Viruses. 2022. PMID: 36146816 Free PMC article. Review.
-
A cluster of transcripts encoded by KSHV ORF30-33 gene locus.Virus Genes. 2012 Apr;44(2):225-36. doi: 10.1007/s11262-011-0698-1. Epub 2011 Dec 17. Virus Genes. 2012. PMID: 22180077 Free PMC article.
-
Replication kinetics of duck enteritis virus UL16 gene in vitro.Virol J. 2012 Nov 21;9:281. doi: 10.1186/1743-422X-9-281. Virol J. 2012. PMID: 23171438 Free PMC article.
-
Liquid-liquid phase separation mediates the formation of herpesvirus assembly compartments.J Cell Biol. 2023 Jan 2;222(1):e202201088. doi: 10.1083/jcb.202201088. Epub 2022 Oct 17. J Cell Biol. 2023. PMID: 36250941 Free PMC article.
-
Complex mechanisms for the packaging of the UL16 tegument protein into herpes simplex virus.Virology. 2010 Mar 15;398(2):208-13. doi: 10.1016/j.virol.2009.12.004. Epub 2010 Jan 3. Virology. 2010. PMID: 20051283 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

