Hepatitis C virus utilizes lipid droplet for production of infectious virus
- PMID: 19644222
- PMCID: PMC3561845
- DOI: 10.2183/pjab.85.217
Hepatitis C virus utilizes lipid droplet for production of infectious virus
Abstract
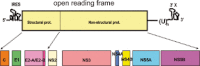
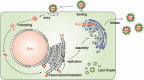
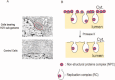
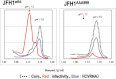
Hepatitis C virus (HCV) establishes a persistent infection and causes chronic hepatitis. Chronic hepatitis patients often develop hepatic cirrhosis and progress to liver cancer. The development of this pathological condition is linked to the persistent infection of the virus. In other words, viral replication/multiplication may contribute to disease pathology. Accumulating clinical studies suggest that HCV infection alters lipid metabolism, and thus causes fatty liver. It has been reported that this abnormal metabolism exacerbates hepatic diseases. Recently, we revealed that lipid droplets play a key role in HCV replication. Understanding the molecular mechanism of HCV replication will help elucidate the pathogenic mechanism and develop preventive measures that inhibit disease manifestation by blocking persistent infection. In this review, we outline recent findings on the function of lipid droplets in the HCV replication cycle and describe the relationship between the development of liver diseases and virus replication.
Figures


Similar articles
-
The Role of Cytosolic Lipid Droplets in Hepatitis C Virus Replication, Assembly, and Release.Biomed Res Int. 2023 Apr 14;2023:5156601. doi: 10.1155/2023/5156601. eCollection 2023. Biomed Res Int. 2023. PMID: 37090186 Free PMC article. Review.
-
N-Myc Downstream-Regulated Gene 1 Restricts Hepatitis C Virus Propagation by Regulating Lipid Droplet Biogenesis and Viral Assembly.J Virol. 2018 Jan 2;92(2):e01166-17. doi: 10.1128/JVI.01166-17. Print 2018 Jan 15. J Virol. 2018. PMID: 29118118 Free PMC article.
-
[Advances in research on HCV replication and virion formation].Uirusu. 2010 Jun;60(1):87-92. doi: 10.2222/jsv.60.87. Uirusu. 2010. PMID: 20848868 Review. Japanese.
-
Complex lipid metabolic remodeling is required for efficient hepatitis C virus replication.Biochim Biophys Acta Mol Cell Biol Lipids. 2018 Sep;1863(9):1041-1056. doi: 10.1016/j.bbalip.2018.06.002. Epub 2018 Jun 6. Biochim Biophys Acta Mol Cell Biol Lipids. 2018. PMID: 29885363
-
USP15 Participates in Hepatitis C Virus Propagation through Regulation of Viral RNA Translation and Lipid Droplet Formation.J Virol. 2019 Mar 5;93(6):e01708-18. doi: 10.1128/JVI.01708-18. Print 2019 Mar 15. J Virol. 2019. PMID: 30626683 Free PMC article.
Cited by
-
Global Lipidome Profiling Revealed Multifaceted Role of Lipid Species in Hepatitis C Virus Replication, Assembly, and Host Antiviral Response.Viruses. 2023 Feb 7;15(2):464. doi: 10.3390/v15020464. Viruses. 2023. PMID: 36851679 Free PMC article.
-
Lipids at the Crossroad of α-Synuclein Function and Dysfunction: Biological and Pathological Implications.Front Cell Neurosci. 2019 May 1;13:175. doi: 10.3389/fncel.2019.00175. eCollection 2019. Front Cell Neurosci. 2019. PMID: 31118888 Free PMC article. Review.
-
Inhibitory effects of bile acids and synthetic farnesoid X receptor agonists on rotavirus replication.J Virol. 2011 Dec;85(23):12570-7. doi: 10.1128/JVI.05839-11. Epub 2011 Sep 28. J Virol. 2011. PMID: 21957312 Free PMC article.
-
Dengue virus-induced ER stress is required for autophagy activation, viral replication, and pathogenesis both in vitro and in vivo.Sci Rep. 2018 Jan 11;8(1):489. doi: 10.1038/s41598-017-18909-3. Sci Rep. 2018. PMID: 29323257 Free PMC article.
-
Prominent steatosis with hypermetabolism of the cell line permissive for years of infection with hepatitis C virus.PLoS One. 2014 Apr 9;9(4):e94460. doi: 10.1371/journal.pone.0094460. eCollection 2014. PLoS One. 2014. PMID: 24718268 Free PMC article.
References
-
- Lindenbach, B.D., Thiel, H.J. and Rice, C. (2007) The viruses and their replication. InField’s Virology (eds. Knipe, D.M. and Howley, P.M.). Lippincott-Raven, Philadelphia, pp. 1101–1152

