Intramembrane glycine mediates multimerization of Insig-2, a requirement for sterol regulation in Chinese hamster ovary cells
- PMID: 19617589
- PMCID: PMC2789779
- DOI: 10.1194/jlr.M900336-JLR200
Intramembrane glycine mediates multimerization of Insig-2, a requirement for sterol regulation in Chinese hamster ovary cells
Abstract
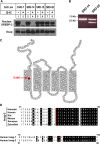
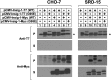
Sterol-induced binding of endoplasmic reticulum (ER) membrane proteins Insig-1 and Insig-2 to SREBP cleavage-activating protein (Scap) and HMG-CoA reductase triggers regulatory events that limit cholesterol synthesis in animal cells. Binding of Insigs to Scap prevents proteolytic activation of sterol-regulatory element binding proteins (SREBPs), membrane-bound transcription factors that enhance cholesterol synthesis, by trapping Scap-SREBP complexes in the ER. Insig binding to reductase causes ubiquitination and subsequent proteasome-mediated degradation of the enzyme from ER membranes, slowing a rate-limiting step in cholesterol synthesis. Here, we report the characterization of mutant Chinese hamster ovary cells, designated SRD-20, that are resistant to 25-hydroxycholesterol, which potently inhibits SREBP activation and stimulates degradation of reductase. SRD-20 cells were produced by mutagenesis of Insig-1-deficient SRD-14 cells, followed by selection in 25-hydroxycholesterol. DNA sequencing reveals that SRD-20 cells harbor a point mutation in one Insig-2 allele that results in production of a truncated, nonfunctional protein, whereas the other allele contains a point mutation that results in substitution of glutamic acid for glycine-39. This glycine residue localizes to the first membrane-spanning segment of Insig-2 and is also present in the corresponding region of Insig-1. Mutant forms of Insig-1 and Insig-2 containing the Glu-to-Gly substitution fail to confer sterol regulation upon overexpressed Scap and reductase. These studies identify the intramembrane glycine as a key residue for normal sterol regulation in animal cells.
Figures



Similar articles
-
Dual functions of Insig proteins in cholesterol homeostasis.Lipids Health Dis. 2012 Dec 18;11:173. doi: 10.1186/1476-511X-11-173. Lipids Health Dis. 2012. PMID: 23249523 Free PMC article. Review.
-
Isolation of sterol-resistant Chinese hamster ovary cells with genetic deficiencies in both Insig-1 and Insig-2.J Biol Chem. 2005 Jul 1;280(26):25242-9. doi: 10.1074/jbc.M502989200. Epub 2005 May 2. J Biol Chem. 2005. PMID: 15866869
-
Amplification of the gene for SCAP, coupled with Insig-1 deficiency, confers sterol resistance in mutant Chinese hamster ovary cells.J Lipid Res. 2007 Sep;48(9):1944-54. doi: 10.1194/jlr.M700225-JLR200. Epub 2007 Jun 22. J Lipid Res. 2007. PMID: 17586788
-
Isolation of mutant cells lacking Insig-1 through selection with SR-12813, an agent that stimulates degradation of 3-hydroxy-3-methylglutaryl-coenzyme A reductase.J Biol Chem. 2004 Oct 8;279(41):43136-47. doi: 10.1074/jbc.M406406200. Epub 2004 Jul 7. J Biol Chem. 2004. PMID: 15247248
-
Underlying mechanisms for sterol-induced ubiquitination and ER-associated degradation of HMG CoA reductase.Semin Cell Dev Biol. 2018 Sep;81:121-128. doi: 10.1016/j.semcdb.2017.10.019. Epub 2017 Nov 7. Semin Cell Dev Biol. 2018. PMID: 29107682 Free PMC article. Review.
Cited by
-
Cell cholesterol homeostasis: mediation by active cholesterol.Trends Cell Biol. 2010 Nov;20(11):680-7. doi: 10.1016/j.tcb.2010.08.007. Epub 2010 Sep 16. Trends Cell Biol. 2010. PMID: 20843692 Free PMC article. Review.
-
RNA-Seq based transcriptome analysis during bovine viral diarrhoea virus (BVDV) infection.BMC Genomics. 2019 Oct 24;20(1):774. doi: 10.1186/s12864-019-6120-4. BMC Genomics. 2019. PMID: 31651237 Free PMC article.
-
Dual functions of Insig proteins in cholesterol homeostasis.Lipids Health Dis. 2012 Dec 18;11:173. doi: 10.1186/1476-511X-11-173. Lipids Health Dis. 2012. PMID: 23249523 Free PMC article. Review.
References
-
- Goldstein J. L., DeBose-Boyd R. A., Brown M. S. 2006. Protein sensors for membrane sterols. Cell. 124: 35–46. - PubMed
-
- Nohturfft A., Yabe D., Goldstein J. L., Brown M. S., Espenshade P. J. 2000. Regulated step in cholesterol feedback localized to budding of SCAP from ER membranes. Cell. 102: 315–323. - PubMed
-
- DeBose-Boyd R. A., Brown M. S., Li W. P., Nohturfft A., Goldstein J. L., Espenshade P. J. 1999. Transport-dependent proteolysis of SREBP: relocation of site-1 protease from Golgi to ER obviates the need for SREBP transport to Golgi. Cell. 99: 703–712. - PubMed
-
- Goldstein J. L., Brown M. S. 1990. Regulation of the mevalonate pathway. Nature. 343: 425–430. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

