Global analysis of alternative splicing uncovers developmental regulation of nonsense-mediated decay in C. elegans
- PMID: 19617316
- PMCID: PMC2743056
- DOI: 10.1261/rna.1711109
Global analysis of alternative splicing uncovers developmental regulation of nonsense-mediated decay in C. elegans
Abstract
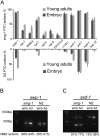
Alternative splicing coupled to nonsense-mediated decay (AS-NMD) is a mechanism for post-transcriptional regulation of gene expression. We analyzed the global effects of mutations in seven genes of the C. elegans NMD pathway on AS isoform ratios. We find that mutations in two NMD factors, smg-6 and smg-7, have weaker global effects relative to mutations in other smg genes. We did an in-depth analysis of 12 pre-mRNA splicing factor genes that are subject to AS-NMD. For four of these, changes in the ratio of alternatively spliced isoforms during development are caused by developmentally regulated inhibition of NMD, and not by changes in alternative splicing. Using sucrose gradient analysis of mRNAs undergoing translation, we find several examples of NMD-dependent enrichment of premature termination codon (PTC) isoforms in the monosome fraction. In contrast, we present evidence of two genes for which the PTC-containing isoforms are found in polysomes and have a translational profile similar to non-PTC-containing transcripts from the same gene. We propose that NMD of certain alternatively spliced isoforms is regulated, and that some stabilized NMD targets may be translated.
Figures

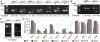
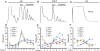

Similar articles
-
Alternative splicing regulation during C. elegans development: splicing factors as regulated targets.PLoS Genet. 2008 Feb 29;4(2):e1000001. doi: 10.1371/journal.pgen.1000001. PLoS Genet. 2008. PMID: 18454200 Free PMC article.
-
Mutations in genes involved in nonsense mediated decay ameliorate the phenotype of sel-12 mutants with amber stop mutations in Caenorhabditis elegans.BMC Genet. 2009 Mar 20;10:14. doi: 10.1186/1471-2156-10-14. BMC Genet. 2009. PMID: 19302704 Free PMC article.
-
The Substrates of Nonsense-Mediated mRNA Decay in Caenorhabditis elegans.G3 (Bethesda). 2018 Jan 4;8(1):195-205. doi: 10.1534/g3.117.300254. G3 (Bethesda). 2018. PMID: 29122854 Free PMC article.
-
The coupling of alternative splicing and nonsense-mediated mRNA decay.Adv Exp Med Biol. 2007;623:190-211. doi: 10.1007/978-0-387-77374-2_12. Adv Exp Med Biol. 2007. PMID: 18380348 Review.
-
Nonsense-mediated RNA decay regulation by cellular stress: implications for tumorigenesis.Mol Cancer Res. 2010 Mar;8(3):295-308. doi: 10.1158/1541-7786.MCR-09-0502. Epub 2010 Feb 23. Mol Cancer Res. 2010. PMID: 20179151 Free PMC article. Review.
Cited by
-
Frac-seq reveals isoform-specific recruitment to polyribosomes.Genome Res. 2013 Oct;23(10):1615-23. doi: 10.1101/gr.148585.112. Epub 2013 Jun 19. Genome Res. 2013. PMID: 23783272 Free PMC article.
-
Alternative Splicing Regulation of Cancer-Related Pathways in Caenorhabditis elegans: An In Vivo Model System with a Powerful Reverse Genetics Toolbox.Int J Cell Biol. 2013;2013:636050. doi: 10.1155/2013/636050. Epub 2013 Aug 28. Int J Cell Biol. 2013. PMID: 24069034 Free PMC article. Review.
-
Nonsense mRNA suppression via nonstop decay.Elife. 2018 Jan 8;7:e33292. doi: 10.7554/eLife.33292. Elife. 2018. PMID: 29309033 Free PMC article.
-
Co-regulation of alternative splicing by diverse splicing factors in Caenorhabditis elegans.Nucleic Acids Res. 2011 Jan;39(2):666-74. doi: 10.1093/nar/gkq767. Epub 2010 Aug 30. Nucleic Acids Res. 2011. PMID: 20805248 Free PMC article.
-
Physiological Consequences of Nonsense-Mediated Decay and Its Role in Adaptive Responses.Biomedicines. 2024 May 16;12(5):1110. doi: 10.3390/biomedicines12051110. Biomedicines. 2024. PMID: 38791071 Free PMC article. Review.
References
-
- Ben-Dov C, Hartmann B, Lundgren J, Valcarcel J. Genome-wide analysis of alternative pre-mRNA splicing. J Biol Chem. 2008;283:1229–1233. - PubMed
-
- Berke JD, Sgambato V, Zhu PP, Lavoie B, Vincent M, Krause M, Hyman SE. Dopamine and glutamate induce distinct striatal splice forms of Ania-6, an RNA polymerase II-associated cyclin. Neuron. 2001;32:277–287. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
