A comparative study of HIV-1 clade C env evolution in a Zambian infant with an infected rhesus macaque during disease progression
- PMID: 19609201
- PMCID: PMC2901162
- DOI: 10.1097/QAD.0b013e32832f3da6
A comparative study of HIV-1 clade C env evolution in a Zambian infant with an infected rhesus macaque during disease progression
Abstract
Objective: To evaluate whether HIV-1 clade C (HIV-C) envelope variations that arise during disease progression in rhesus macaque model reflect changes that occur naturally in human infection.
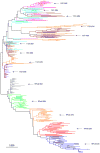
Design: An infant macaque was infected with SHIV-1157i, an R5 tropic clade C SHIV, that expresses a primary HIV-C envelope derived from an infected human infant and monitored over a 5-year period. Genetic variation of the V1-V5 envelope region, which is the main target for humoral immune responses, derived from the infected macaque and infant was examined.
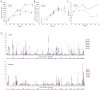
Methods: The V1-V5 envelope region was cloned and sequenced from longitudinal peripheral blood mononuclear cell samples collected from the infected macaque and infant. Phylogenetic analysis [phylogenetic tree, diversity, divergence, ratio of nonsynonymous (dN) and synonymous substitution (dS) and dN distribution] was performed. Plasma RNA viral load, CD4(+) T-cell count, changes in the length of V1-V5 region, putative N-linked glycosylation site number and distribution were also measured.
Results: Phylogenetic analysis revealed that changes in the macaque closely reflected those of the infant during disease progression. Similar distribution patterns of dN and hot spots were observed between the macaque and infant. Analysis of putative N-linked glycosylation sites revealed several common variations between the virus populations in the two host species. These variations correlate with decline of CD4 T-cell count in the macaque and might be linked with disease progression.
Conclusion: SHIV-C infection of macaque is a relevant animal model for studying variation of primary HIV-C envelope during disease progression and could be used to analyze the selection pressures that are associated with those changes.
2009 Wolters Kluwer Health | Lippincott Williams & Wilkins
Figures

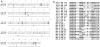
Similar articles
-
Development of a novel rhesus macaque model with an infectious R5 simian-human immunodeficiency virus encoding HIV-1 CRF08_BC env.J Med Primatol. 2014 Feb;43(1):11-21. doi: 10.1111/jmp.12084. Epub 2013 Sep 11. J Med Primatol. 2014. PMID: 24020838
-
Molecularly cloned SHIV-1157ipd3N4: a highly replication- competent, mucosally transmissible R5 simian-human immunodeficiency virus encoding HIV clade C Env.J Virol. 2006 Sep;80(17):8729-38. doi: 10.1128/JVI.00558-06. J Virol. 2006. PMID: 16912320 Free PMC article.
-
SHIV-1157i and passaged progeny viruses encoding R5 HIV-1 clade C env cause AIDS in rhesus monkeys.Retrovirology. 2008 Oct 17;5:94. doi: 10.1186/1742-4690-5-94. Retrovirology. 2008. PMID: 18928523 Free PMC article.
-
CD4-HIV-1 Envelope Interactions: Critical Insights for the Simian/HIV/Macaque Model.AIDS Res Hum Retroviruses. 2018 Sep;34(9):778-779. doi: 10.1089/AID.2018.0110. Epub 2018 Jul 9. AIDS Res Hum Retroviruses. 2018. PMID: 29886767 Free PMC article. Review.
-
Understanding the basis of CD4(+) T-cell depletion in macaques infected by a simian-human immunodeficiency virus.Vaccine. 2002 May 6;20(15):1934-7. doi: 10.1016/s0264-410x(02)00072-5. Vaccine. 2002. PMID: 11983249 Review.
Cited by
-
Coreceptor use in nonhuman primate models of HIV infection.J Transl Med. 2011 Jan 27;9 Suppl 1(Suppl 1):S7. doi: 10.1186/1479-5876-9-S1-S7. J Transl Med. 2011. PMID: 21284906 Free PMC article. Review.
-
Characterization of a new simian immunodeficiency virus strain in a naturally infected Pan troglodytes troglodytes chimpanzee with AIDS related symptoms.Retrovirology. 2011 Jan 13;8:4. doi: 10.1186/1742-4690-8-4. Retrovirology. 2011. PMID: 21232091 Free PMC article.
-
Variations in the Biological Functions of HIV-1 Clade C Envelope in a SHIV-Infected Rhesus Macaque during Disease Progression.PLoS One. 2013 Jun 26;8(6):e66973. doi: 10.1371/journal.pone.0066973. Print 2013. PLoS One. 2013. PMID: 23840566 Free PMC article.
-
Restricted genetic diversity of HIV-1 subtype C envelope glycoprotein from perinatally infected Zambian infants.PLoS One. 2010 Feb 18;5(2):e9294. doi: 10.1371/journal.pone.0009294. PLoS One. 2010. PMID: 20174636 Free PMC article.
-
HIV-1 Env C2-V4 diversification in a slow-progressor infant reveals a flat but rugged fitness landscape.PLoS One. 2013 Apr 29;8(4):e63094. doi: 10.1371/journal.pone.0063094. Print 2013. PLoS One. 2013. PMID: 23638182 Free PMC article.
References
-
- Gaschen B, Taylor J, Yusim K, Foley B, Gao F, Lang D, et al. Diversity considerations in HIV-1 vaccine selection. Science. 2002;296:2354–2360. - PubMed
-
- Geyer H, Holschbach C, Hunsmann G, Schneider J. Carbohydrates of human immunodeficiency virus. Structures of oligosaccharides linked to the envelope glycoprotein 120. J Biol Chem. 1988;263:11760–11767. - PubMed
-
- Stevceva L, Yoon V, Anastasiades D, Poznansky MC. Immune responses to HIV Gp120 that facilitate viral escape. Curr HIV Res. 2007;5:47–54. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

