Toxoplasma gondii cathepsin L is the primary target of the invasion-inhibitory compound morpholinurea-leucyl-homophenyl-vinyl sulfone phenyl
- PMID: 19596863
- PMCID: PMC2785372
- DOI: 10.1074/jbc.M109.003780
Toxoplasma gondii cathepsin L is the primary target of the invasion-inhibitory compound morpholinurea-leucyl-homophenyl-vinyl sulfone phenyl
Abstract
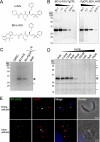
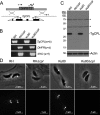
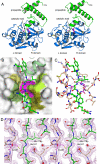
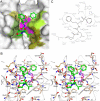
The protozoan parasite Toxoplasma gondii relies on post-translational modification, including proteolysis, of proteins required for recognition and invasion of host cells. We have characterized the T. gondii cysteine protease cathepsin L (TgCPL), one of five cathepsins found in the T. gondii genome. We show that TgCPL is the primary target of the compound morpholinurea-leucyl-homophenyl-vinyl sulfone phenyl (LHVS), which was previously shown to inhibit parasite invasion by blocking the release of invasion proteins from microneme secretory organelles. As shown by fluorescently labeled LHVS and TgCPL-specific antibodies, TgCPL is associated with a discrete vesicular structure in the apical region of extracellular parasites but is found in multiple puncta throughout the cytoplasm of intracellular replicating parasites. LHVS fails to label cells lacking TgCPL due to targeted disruption of the TgCPL gene in two different parasite strains. We present a structural model for the inhibition of TgCPL by LHVS based on a 2.0 A resolution crystal structure of TgCPL in complex with its propeptide. We discuss possible roles for TgCPL as a protease involved in the degradation or limited proteolysis of parasite proteins involved in invasion.
Figures

Similar articles
-
The cathepsin L of Toxoplasma gondii (TgCPL) and its endogenous macromolecular inhibitor, toxostatin.Mol Biochem Parasitol. 2009 Mar;164(1):86-94. doi: 10.1016/j.molbiopara.2008.11.012. Epub 2008 Dec 6. Mol Biochem Parasitol. 2009. PMID: 19111576 Free PMC article.
-
Determination of Chemical Inhibitor Efficiency against Intracellular Toxoplasma Gondii Growth Using a Luciferase-Based Growth Assay.J Vis Exp. 2020 Apr 29;(158):10.3791/60985. doi: 10.3791/60985. J Vis Exp. 2020. PMID: 32420988 Free PMC article.
-
Toxoplasma gondii cathepsin proteases are undeveloped prominent vaccine antigens against toxoplasmosis.BMC Infect Dis. 2013 May 7;13:207. doi: 10.1186/1471-2334-13-207. BMC Infect Dis. 2013. PMID: 23651838 Free PMC article.
-
Cathepsin proteases in Toxoplasma gondii.Adv Exp Med Biol. 2011;712:49-61. doi: 10.1007/978-1-4419-8414-2_4. Adv Exp Med Biol. 2011. PMID: 21660658 Free PMC article. Review.
-
Cruzain : the path from target validation to the clinic.Adv Exp Med Biol. 2011;712:100-15. doi: 10.1007/978-1-4419-8414-2_7. Adv Exp Med Biol. 2011. PMID: 21660661 Review.
Cited by
-
Covalent Reversible Inhibitors of Cysteine Proteases Containing the Nitrile Warhead: Recent Advancement in the Field of Viral and Parasitic Diseases.Molecules. 2022 Apr 15;27(8):2561. doi: 10.3390/molecules27082561. Molecules. 2022. PMID: 35458759 Free PMC article. Review.
-
Optimization of dipeptidic inhibitors of cathepsin L for improved Toxoplasma gondii selectivity and CNS permeability.Bioorg Med Chem Lett. 2018 Jun 1;28(10):1972-1980. doi: 10.1016/j.bmcl.2018.03.020. Epub 2018 Mar 9. Bioorg Med Chem Lett. 2018. PMID: 29650289 Free PMC article.
-
Toxoplasma Cathepsin Protease B and Aspartyl Protease 1 Are Dispensable for Endolysosomal Protein Digestion.mSphere. 2020 Feb 12;5(1):e00869-19. doi: 10.1128/mSphere.00869-19. mSphere. 2020. PMID: 32051238 Free PMC article.
-
Role of Toxoplasma gondii Chloroquine Resistance Transporter in Bradyzoite Viability and Digestive Vacuole Maintenance.mBio. 2019 Aug 6;10(4):e01324-19. doi: 10.1128/mBio.01324-19. mBio. 2019. PMID: 31387907 Free PMC article.
-
Novel N-benzoyl-2-hydroxybenzamide disrupts unique parasite secretory pathway.Antimicrob Agents Chemother. 2012 May;56(5):2666-82. doi: 10.1128/AAC.06450-11. Epub 2012 Feb 21. Antimicrob Agents Chemother. 2012. PMID: 22354304 Free PMC article.
References
-
- Puente X. S., Sánchez L. M., Overall C. M., López-Otín C. (2003) Nat. Rev. Genet. 4, 544–558 - PubMed
-
- Rosenthal P. J. (2004) Int. J. Parasitol. 34, 1489–1499 - PubMed
-
- McKerrow J. H., Caffrey C., Kelly B., Loke P., Sajid M. (2006) Annu. Rev. Pathol. 1, 497–536 - PubMed
-
- Keeley A., Soldati D. (2004) Trends Cell Biol. 14, 528–532 - PubMed
-
- Carruthers V., Boothroyd J. C. (2007) Curr. Opin. Microbiol. 10, 83–89 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

