Yeast two-hybrid, a powerful tool for systems biology
- PMID: 19582228
- PMCID: PMC2705515
- DOI: 10.3390/ijms10062763
Yeast two-hybrid, a powerful tool for systems biology
Abstract
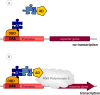
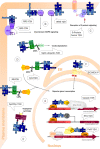
A key property of complex biological systems is the presence of interaction networks formed by its different components, primarily proteins. These are crucial for all levels of cellular function, including architecture, metabolism and signalling, as well as the availability of cellular energy. Very stable, but also rather transient and dynamic protein-protein interactions generate new system properties at the level of multiprotein complexes, cellular compartments or the entire cell. Thus, interactomics is expected to largely contribute to emerging fields like systems biology or systems bioenergetics. The more recent technological development of high-throughput methods for interactomics research will dramatically increase our knowledge of protein interaction networks. The two most frequently used methods are yeast two-hybrid (Y2H) screening, a well established genetic in vivo approach, and affinity purification of complexes followed by mass spectrometry analysis, an emerging biochemical in vitro technique. So far, a majority of published interactions have been detected using an Y2H screen. However, with the massive application of this method, also some limitations have become apparent. This review provides an overview on available yeast two-hybrid methods, in particular focusing on more recent approaches. These allow detection of protein interactions in their native environment, as e.g. in the cytosol or bound to a membrane, by using cytosolic signalling cascades or split protein constructs. Strengths and weaknesses of these genetic methods are discussed and some guidelines for verification of detected protein-protein interactions are provided.
Keywords: interactomics; mass spectrometry; protein-protein interaction; systems bioenergetics; yeast two-hybrid.
Figures
Similar articles
-
Macromolecular crowding: chemistry and physics meet biology (Ascona, Switzerland, 10-14 June 2012).Phys Biol. 2013 Aug;10(4):040301. doi: 10.1088/1478-3975/10/4/040301. Epub 2013 Aug 2. Phys Biol. 2013. PMID: 23912807
-
Mapping Protein-Protein Interaction Using High-Throughput Yeast 2-Hybrid.Methods Mol Biol. 2017;1610:217-230. doi: 10.1007/978-1-4939-7003-2_14. Methods Mol Biol. 2017. PMID: 28439866
-
The yeast two-hybrid system: a tool for mapping protein-protein interactions.Cold Spring Harb Protoc. 2015 May 1;2015(5):425-30. doi: 10.1101/pdb.top083345. Cold Spring Harb Protoc. 2015. PMID: 25934943
-
Adding biological meaning to human protein-protein interactions identified by yeast two-hybrid screenings: A guide through bioinformatics tools.J Proteomics. 2018 Jan 16;171:127-140. doi: 10.1016/j.jprot.2017.05.012. Epub 2017 May 16. J Proteomics. 2018. PMID: 28526529 Review.
-
Translational Metabolomics of Head Injury: Exploring Dysfunctional Cerebral Metabolism with Ex Vivo NMR Spectroscopy-Based Metabolite Quantification.In: Kobeissy FH, editor. Brain Neurotrauma: Molecular, Neuropsychological, and Rehabilitation Aspects. Boca Raton (FL): CRC Press/Taylor & Francis; 2015. Chapter 25. In: Kobeissy FH, editor. Brain Neurotrauma: Molecular, Neuropsychological, and Rehabilitation Aspects. Boca Raton (FL): CRC Press/Taylor & Francis; 2015. Chapter 25. PMID: 26269925 Free Books & Documents. Review.
Cited by
-
AKIN10 delays flowering by inactivating IDD8 transcription factor through protein phosphorylation in Arabidopsis.BMC Plant Biol. 2015 May 1;15:110. doi: 10.1186/s12870-015-0503-8. BMC Plant Biol. 2015. PMID: 25929516 Free PMC article.
-
Proteomic approaches to investigate gammaherpesvirus biology and associated tumorigenesis.Adv Virus Res. 2021;109:201-254. doi: 10.1016/bs.aivir.2020.10.001. Epub 2020 Nov 9. Adv Virus Res. 2021. PMID: 33934828 Free PMC article. Review.
-
Keeping in Touch with Type-III Secretion System Effectors: Mass Spectrometry-Based Proteomics to Study Effector-Host Protein-Protein Interactions.Int J Mol Sci. 2020 Sep 19;21(18):6891. doi: 10.3390/ijms21186891. Int J Mol Sci. 2020. PMID: 32961832 Free PMC article. Review.
-
MyProteinNet: build up-to-date protein interaction networks for organisms, tissues and user-defined contexts.Nucleic Acids Res. 2015 Jul 1;43(W1):W258-63. doi: 10.1093/nar/gkv515. Epub 2015 May 18. Nucleic Acids Res. 2015. PMID: 25990735 Free PMC article.
-
The Wheat GENIE3 Network Provides Biologically-Relevant Information in Polyploid Wheat.G3 (Bethesda). 2020 Oct 5;10(10):3675-3686. doi: 10.1534/g3.120.401436. G3 (Bethesda). 2020. PMID: 32747342 Free PMC article.
References
-
- Kiemer L, Cesareni G. Comparative interactomics: comparing apples and pears? Trends Biotechnol. 2007;25:448–454. - PubMed
-
- Kelly W, Stumpf M. Protein-protein interactions: from global to local analyses. Curr. Opin. Biotechnol. 2008;19:396–403. - PubMed
-
- Charbonnier S, Gallego O, Gavin AC. The social network of a cell: recent advances in interactome mapping. Biotechnol. Annu. Rev. 2008;14:1–28. - PubMed
-
- Scheffler IE. Mitochondria make a come back. Adv. Drug. Deliv. Rev. 2001;49:3–26. - PubMed
-
- Scheffler IE. Mitochondria. 2nd ed. John Wiley & Sons; Hoboken, New Jersey, USA: 2007.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

