Determination of quantitative and site-specific DNA methylation of perforin by pyrosequencing
- PMID: 19523225
- PMCID: PMC2704226
- DOI: 10.1186/1756-0500-2-104
Determination of quantitative and site-specific DNA methylation of perforin by pyrosequencing
Abstract
Background: Differential expression of perforin (PRF1), a gene with a pivotal role in immune surveillance, can be attributed to differential methylation of CpG sites in its promoter region. A reproducible method for quantitative and CpG site-specific determination of perforin methylation is required for molecular epidemiologic studies of chronic diseases with immune dysfunction.
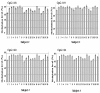
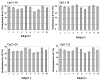
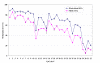
Findings: We developed a pyrosequencing based method to quantify site-specific methylation levels in 32 out of 34 CpG sites in the PRF1 promoter, and also compared methylation pattern in DNAs extracted from whole blood drawn into PAXgene blood DNA tubes (whole blood DNA) or DNA extracted from peripheral blood mononuclear cells (PBMC DNA) from the same normal subjects. Sodium bisulfite treatment of DNA and touchdown PCR were highly reproducible (coefficient of variation 1.63 to 2.18%) to preserve methylation information. Application of optimized pyrosequencing protocol to whole blood DNA revealed that methylation level varied along the promoter in normal subjects with extremely high methylation (mean 86%; range 82-92%) in the distal enhancer region (CpG sites 1-10), a variable methylation (range 49%-83%) in the methylation sensitive region (CpG sites 11-17), and a progressively declining methylation level (range 12%-80%) in the proximal promoter region (CpG sites 18-32) of PRF1. This pattern of methylation remained the same between whole blood and PBMC DNAs, but the absolute values of methylation in 30 out of 32 CpG sites differed significantly, with higher values for all CpG sites in the whole blood DNA.
Conclusion: This reproducible, site-specific and quantitative method for methylation determination of PRF1 based on pyrosequencing without cloning is well suited for large-scale molecular epidemiologic studies of diseases with immune dysfunction. PBMC DNA may be better suited than whole blood DNA for examining methylation levels in genes associated with immune function.
Figures
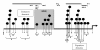
Similar articles
-
Cord blood PRF1 methylation patterns and risk of lower respiratory tract infections in infants: findings from the Ulm Birth Cohort.Medicine (Baltimore). 2015 Jan;94(1):e332. doi: 10.1097/MD.0000000000000332. Medicine (Baltimore). 2015. PMID: 25569648 Free PMC article.
-
Acute psychosocial stress-mediated changes in the expression and methylation of perforin in chronic fatigue syndrome.Genet Epigenet. 2013 Jan 28;5:1-9. doi: 10.4137/GEG.S10944. eCollection 2013. Genet Epigenet. 2013. PMID: 25512702 Free PMC article.
-
Reduced PRF1 enhancer methylation in children with a history of severe RSV bronchiolitis in infancy: an association study.BMC Pediatr. 2017 Mar 3;17(1):65. doi: 10.1186/s12887-017-0817-9. BMC Pediatr. 2017. PMID: 28253869 Free PMC article.
-
Pyrosequencing assays to study promoter CpG site methylation of the O6-MGMT, hMLH1, p14ARF, p16INK4a, RASSF1A, and APC1A genes.Oncol Rep. 2009 Mar;21(3):721-9. Oncol Rep. 2009. PMID: 19212632
-
Quantitation of site-specific HPV 16 DNA methylation by pyrosequencing.J Virol Methods. 2006 Dec;138(1-2):170-6. doi: 10.1016/j.jviromet.2006.08.012. Epub 2006 Oct 11. J Virol Methods. 2006. PMID: 17045346
Cited by
-
The transcription factors Runx3 and ThPOK cross-regulate acquisition of cytotoxic function by human Th1 lymphocytes.Elife. 2018 Feb 28;7:e30496. doi: 10.7554/eLife.30496. Elife. 2018. PMID: 29488879 Free PMC article.
-
DNA methylation within the I.4 promoter region correlates with CYPl19A1 gene expression in human ex vivo mature omental and subcutaneous adipocytes.BMC Med Genet. 2013 Aug 30;14:87. doi: 10.1186/1471-2350-14-87. BMC Med Genet. 2013. PMID: 24128150 Free PMC article.
-
Cord blood PRF1 methylation patterns and risk of lower respiratory tract infections in infants: findings from the Ulm Birth Cohort.Medicine (Baltimore). 2015 Jan;94(1):e332. doi: 10.1097/MD.0000000000000332. Medicine (Baltimore). 2015. PMID: 25569648 Free PMC article.
-
CpG Methylation Analysis of HPV16 in Laser Capture Microdissected Archival Tissue and Whole Tissue Sections from High Grade Anal Squamous Intraepithelial Lesions: A Potential Disease Biomarker.PLoS One. 2016 Aug 16;11(8):e0160673. doi: 10.1371/journal.pone.0160673. eCollection 2016. PLoS One. 2016. PMID: 27529629 Free PMC article.
-
DNA methylation dynamics in blood after hematopoietic cell transplant.PLoS One. 2013;8(2):e56931. doi: 10.1371/journal.pone.0056931. Epub 2013 Feb 22. PLoS One. 2013. PMID: 23451113 Free PMC article.
References
-
- Gulan G, Ravlic-Gulan J, Strbo N, Sotosek V, Nemec B, Matovinovic D, et al. Systemic and local expression of perforin in lymphocyte subsets in acute and chronic rheumatoid arthritis. J Rheumatol. 2003;30:660–670. - PubMed
-
- Kaplan MJ, Lu Q, Wu A, Attwood J, Richardson B. Demethylation of promoter regulatory elements contributes to perforin overexpression in CD4+ lupus T cells. J Immunol. 2004;172:3652–3661. - PubMed
-
- Lu Q, Wu A, Ray D, Deng C, Attwood J, Hanash S, et al. DNA methylation and chromatin structure regulate T cell perforin gene expression. J Immunol. 2003;170:5124–5132. - PubMed
LinkOut - more resources
Full Text Sources

