GAGE: generally applicable gene set enrichment for pathway analysis
- PMID: 19473525
- PMCID: PMC2696452
- DOI: 10.1186/1471-2105-10-161
GAGE: generally applicable gene set enrichment for pathway analysis
Abstract
Background: Gene set analysis (GSA) is a widely used strategy for gene expression data analysis based on pathway knowledge. GSA focuses on sets of related genes and has established major advantages over individual gene analyses, including greater robustness, sensitivity and biological relevance. However, previous GSA methods have limited usage as they cannot handle datasets of different sample sizes or experimental designs.
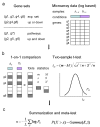
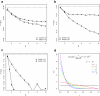
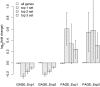
Results: To address these limitations, we present a new GSA method called Generally Applicable Gene-set Enrichment (GAGE). We successfully apply GAGE to multiple microarray datasets with different sample sizes, experimental designs and profiling techniques. GAGE shows significantly better results when compared to two other commonly used GSA methods of GSEA and PAGE. We demonstrate this improvement in the following three aspects: (1) consistency across repeated studies/experiments; (2) sensitivity and specificity; (3) biological relevance of the regulatory mechanisms inferred.GAGE reveals novel and relevant regulatory mechanisms from both published and previously unpublished microarray studies. From two published lung cancer data sets, GAGE derived a more cohesive and predictive mechanistic scheme underlying lung cancer progress and metastasis. For a previously unpublished BMP6 study, GAGE predicted novel regulatory mechanisms for BMP6 induced osteoblast differentiation, including the canonical BMP-TGF beta signaling, JAK-STAT signaling, Wnt signaling, and estrogen signaling pathways-all of which are supported by the experimental literature.
Conclusion: GAGE is generally applicable to gene expression datasets with different sample sizes and experimental designs. GAGE consistently outperformed two most frequently used GSA methods and inferred statistically and biologically more relevant regulatory pathways. The GAGE method is implemented in R in the "gage" package, available under the GNU GPL from http://sysbio.engin.umich.edu/~luow/downloads.php.
Figures


Similar articles
-
Time series gene expression profiling and temporal regulatory pathway analysis of BMP6 induced osteoblast differentiation and mineralization.BMC Syst Biol. 2011 May 23;5:82. doi: 10.1186/1752-0509-5-82. BMC Syst Biol. 2011. PMID: 21605425 Free PMC article.
-
Improving gene set analysis of microarray data by SAM-GS.BMC Bioinformatics. 2007 Jul 5;8:242. doi: 10.1186/1471-2105-8-242. BMC Bioinformatics. 2007. PMID: 17612399 Free PMC article.
-
Learning transcriptional regulatory networks from high throughput gene expression data using continuous three-way mutual information.BMC Bioinformatics. 2008 Nov 3;9:467. doi: 10.1186/1471-2105-9-467. BMC Bioinformatics. 2008. PMID: 18980677 Free PMC article.
-
Investigating the effect of paralogs on microarray gene-set analysis.BMC Bioinformatics. 2011 Jan 24;12:29. doi: 10.1186/1471-2105-12-29. BMC Bioinformatics. 2011. PMID: 21261946 Free PMC article.
-
Concordant integrative gene set enrichment analysis of multiple large-scale two-sample expression data sets.BMC Genomics. 2014;15 Suppl 1(Suppl 1):S6. doi: 10.1186/1471-2164-15-S1-S6. Epub 2014 Jan 24. BMC Genomics. 2014. PMID: 24564564 Free PMC article.
Cited by
-
Exogenous glycogen utilization effects the transcriptome and pathogenicity of Streptococcus suis serotype 2.Front Cell Infect Microbiol. 2022 Nov 9;12:938286. doi: 10.3389/fcimb.2022.938286. eCollection 2022. Front Cell Infect Microbiol. 2022. PMID: 36439226 Free PMC article.
-
Regulation and gene expression profiling of NKG2D positive human cytomegalovirus-primed CD4+ T-cells.PLoS One. 2012;7(8):e41577. doi: 10.1371/journal.pone.0041577. Epub 2012 Aug 1. PLoS One. 2012. PMID: 22870231 Free PMC article.
-
Biomarkers in Liquid Biopsies for Prediction of Early Liver Metastases in Pancreatic Cancer.Cancers (Basel). 2022 Sep 22;14(19):4605. doi: 10.3390/cancers14194605. Cancers (Basel). 2022. PMID: 36230528 Free PMC article.
-
Assessment method for a power analysis to identify differentially expressed pathways.PLoS One. 2012;7(5):e37510. doi: 10.1371/journal.pone.0037510. Epub 2012 May 18. PLoS One. 2012. PMID: 22629411 Free PMC article.
-
Sleep deficits and cannabis use behaviors: an analysis of shared genetics using linkage disequilibrium score regression and polygenic risk prediction.Sleep. 2021 Mar 12;44(3):zsaa188. doi: 10.1093/sleep/zsaa188. Sleep. 2021. PMID: 32935850 Free PMC article.
References
-
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–50. doi: 10.1073/pnas.0506580102. - DOI - PMC - PubMed
-
- Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J, Puigserver P, Carlsson E, Ridderstrale M, Laurila E, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34:267–73. doi: 10.1038/ng1180. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

