On the mechanism of autoinhibition of the RhoA-specific nucleotide exchange factor PDZRhoGEF
- PMID: 19460155
- PMCID: PMC2695464
- DOI: 10.1186/1472-6807-9-36
On the mechanism of autoinhibition of the RhoA-specific nucleotide exchange factor PDZRhoGEF
Abstract
Background: The Dbl-family of guanine nucleotide exchange factors (GEFs) activate the cytosolic GTPases of the Rho family by enhancing the rate of exchange of GTP for GDP on the cognate GTPase. This catalytic activity resides in the DH (Dbl-homology) domain, but typically GEFs are multidomain proteins containing other modules. It is believed that GEFs are autoinhibited in the cytosol due to supramodular architecture, and become activated in diverse signaling pathways through conformational change and exposure of the DH domain, as the protein is translocated to the membrane. A small family of RhoA-specific GEFs, containing the RGSL (regulators of G-protein signaling-like) domain, act as effectors of select GPCRs via Galpha12/13, although the molecular mechanism by which this pathway operates is not known. These GEFs include p115, LARG and PDZRhoGEF (PRG).
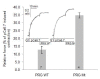
Results: Here we show that the autoinhibition of PRG is caused largely by an interaction of a short negatively charged sequence motif, immediately upstream of the DH-domain and including residues Asp706, Glu708, Glu710 and Asp712, with a patch on the catalytic surface of the DH-domain including Arg867 and Arg868. In the absence of both PDZ and RGSL domains, the DH-PH tandem with additional 21 residues upstream, is 50% autoinhibited. However, within the full-length protein, the PDZ and/or RGSL domains significantly restore autoinhibition.
Conclusion: Our results suggest a mechanism for autoinhibition of RGSL family of GEFs, in which the RGSL domain and a unique sequence motif upstream of the DH domain, act cooperatively to reduce the ability of the DH domain to bind the nucleotide free RhoA. The activation mechanism is likely to involve two independent steps, i.e. displacement of the RGSL domain and conformational change involving the autoinhibitory sequence motif containing several negatively charged residues.
Figures


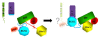
Similar articles
-
Insights into the molecular activation mechanism of the RhoA-specific guanine nucleotide exchange factor, PDZRhoGEF.J Biol Chem. 2011 Oct 7;286(40):35163-75. doi: 10.1074/jbc.M111.270918. Epub 2011 Aug 4. J Biol Chem. 2011. PMID: 21816819 Free PMC article.
-
Mechanistic insights into specificity, activity, and regulatory elements of the regulator of G-protein signaling (RGS)-containing Rho-specific guanine nucleotide exchange factors (GEFs) p115, PDZ-RhoGEF (PRG), and leukemia-associated RhoGEF (LARG).J Biol Chem. 2011 May 20;286(20):18202-12. doi: 10.1074/jbc.M111.226431. Epub 2011 Mar 28. J Biol Chem. 2011. PMID: 21454492 Free PMC article.
-
Real-time NMR study of guanine nucleotide exchange and activation of RhoA by PDZ-RhoGEF.J Biol Chem. 2010 Feb 19;285(8):5137-45. doi: 10.1074/jbc.M109.064691. Epub 2009 Dec 17. J Biol Chem. 2010. PMID: 20018869 Free PMC article.
-
The guanine nucleotide exchange factor Tiam1: a Janus-faced molecule in cellular signaling.Cell Signal. 2014 Mar;26(3):483-91. doi: 10.1016/j.cellsig.2013.11.034. Epub 2013 Dec 2. Cell Signal. 2014. PMID: 24308970 Review.
-
Signaling to the Rho GTPases: networking with the DH domain.FEBS Lett. 2002 Feb 20;513(1):85-91. doi: 10.1016/s0014-5793(01)03310-5. FEBS Lett. 2002. PMID: 11911885 Review.
Cited by
-
C3 transferase gene therapy for continuous conditional RhoA inhibition.Neuroscience. 2016 Dec 17;339:308-318. doi: 10.1016/j.neuroscience.2016.10.022. Epub 2016 Oct 13. Neuroscience. 2016. PMID: 27746349 Free PMC article.
-
Structural determinants of RGS-RhoGEF signaling critical to Entamoeba histolytica pathogenesis.Structure. 2013 Jan 8;21(1):65-75. doi: 10.1016/j.str.2012.11.012. Epub 2012 Dec 20. Structure. 2013. PMID: 23260656 Free PMC article.
-
Semaphorin 4D signaling requires the recruitment of phospholipase C gamma into the plexin-B1 receptor complex.Mol Cell Biol. 2009 Dec;29(23):6321-34. doi: 10.1128/MCB.00103-09. Epub 2009 Oct 5. Mol Cell Biol. 2009. PMID: 19805522 Free PMC article.
-
The role of actin filament dynamics in the myogenic response of cerebral resistance arteries.J Cereb Blood Flow Metab. 2013 Jan;33(1):1-12. doi: 10.1038/jcbfm.2012.144. Epub 2012 Oct 17. J Cereb Blood Flow Metab. 2013. PMID: 23072746 Free PMC article. Review.
-
Activation of p115-RhoGEF requires direct association of Gα13 and the Dbl homology domain.J Biol Chem. 2012 Jul 20;287(30):25490-500. doi: 10.1074/jbc.M111.333716. Epub 2012 Jun 1. J Biol Chem. 2012. PMID: 22661716 Free PMC article.
References
-
- Jaffe AB, Hall A. RHO GTPASES: Biochemistry and Biology. Annu Rev Cell Dev Biol. 2005;21:247–269. - PubMed
-
- Raftopoulou M, Hall A. Cell migration: Rho GTPases lead the way. Dev Biol. 2004;265:23–32. - PubMed
-
- Etienne-Manneville S, Hall A. Rho GTPases in cell biology. Nature. 2002;420:629–635. - PubMed
-
- Kandpal RP. Rho GTPase activating proteins in cancer phenotypes. Curr Protein Pept Sci. 2006;7:355–365. - PubMed
-
- Rossman KL, Der CJ, Sondek J. GEF means go: turning on RHO GTPases with guanine nucleotide-exchange factors. Nat Rev Mol Cell Biol. 2005;6:167–180. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

