Redox-dependent dynamics of a dual thioredoxin fold protein: evolution of specialized folds
- PMID: 19459661
- PMCID: PMC2744581
- DOI: 10.1021/bi900270w
Redox-dependent dynamics of a dual thioredoxin fold protein: evolution of specialized folds
Abstract
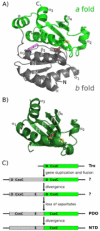
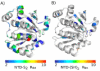
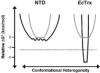
An enzyme system protecting bacteria from oxidative stress includes the flavoprotein AhpF and the peroxiredoxin AhpC. The N-terminal domain of AhpF (NTD), with two fused thioredoxin (Trx) folds, belongs to the hyperthermophilic protein disulfide oxidoreductase family. The NTD is distinct in that it contains a redox active a fold with a CxxC sequence and a redox inactive b fold that has lost the CxxC motif. Here we characterize the stability, the (15)N backbone relaxation, and the hydrogen-deuterium exchange properties of reduced [NTD-(SH)(2)] and oxidized (NTD-S(2)) NTD from Salmonella typhimurium. While both NTD-(SH)(2) and NTD-S(2) exhibit similar equilibrium unfolding transitions and order parameters, R(ex) relaxation terms are quite distinct with considerably more intermediate time scale motions in NTD-S(2). Hydrogen exchange protection factors show that the slowly exchanging core corresponds to residues in the b fold in both NTD-(SH)(2) and NTD-S(2). Interestingly, folded-state dynamic fluctuations in the catalytic a fold are significantly increased for residues in NTD-S(2) compared to NTD-(SH)(2). Taken together, these data demonstrate that oxidation of the active site disulfide does not significantly increase stability but results in a dramatic increase in conformational heterogeneity in residues primarily in the redox active a fold. Differences in dynamics between the two folds of the NTD suggest that each evolved a specialized function which, in the a fold, couples redox state to internal motions which may enhance catalysis and specificity and, in the b fold, provides a redox insensitive stable core.
Figures

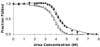




Similar articles
-
Structure of intact AhpF reveals a mirrored thioredoxin-like active site and implies large domain rotations during catalysis.Biochemistry. 2001 Apr 3;40(13):3900-11. doi: 10.1021/bi002765p. Biochemistry. 2001. PMID: 11300769
-
Attachment of the N-terminal domain of Salmonella typhimurium AhpF to Escherichia coli thioredoxin reductase confers AhpC reductase activity but does not affect thioredoxin reductase activity.Biochemistry. 2000 Aug 1;39(30):8859-69. doi: 10.1021/bi000826d. Biochemistry. 2000. PMID: 10913298
-
Essential role of the flexible linker on the conformational equilibrium of bacterial peroxiredoxin reductase for effective regeneration of peroxiredoxin.J Biol Chem. 2017 Apr 21;292(16):6667-6679. doi: 10.1074/jbc.M117.775858. Epub 2017 Mar 7. J Biol Chem. 2017. PMID: 28270505 Free PMC article.
-
Thioredoxin from Escherichia coli as a Role Model of Molecular Recognition, Folding, Dynamics and Function.Protein Pept Lett. 2015;22(9):801-15. doi: 10.2174/0929866522666150707114309. Protein Pept Lett. 2015. PMID: 26149400 Review.
-
Thioredoxins and glutaredoxins as facilitators of protein folding.Biochim Biophys Acta. 2008 Apr;1783(4):641-50. doi: 10.1016/j.bbamcr.2008.02.003. Epub 2008 Feb 19. Biochim Biophys Acta. 2008. PMID: 18331844 Review.
Cited by
-
Peroxiredoxins in parasites.Antioxid Redox Signal. 2012 Aug 15;17(4):608-33. doi: 10.1089/ars.2011.4404. Epub 2012 Jan 25. Antioxid Redox Signal. 2012. PMID: 22098136 Free PMC article. Review.
-
Rational design of multiple TB antigens TB10.4 and TB10.4-Ag85B as subunit vaccine candidates against Mycobacterium tuberculosis.Pharm Res. 2010 Feb;27(2):224-34. doi: 10.1007/s11095-009-9995-y. Epub 2009 Oct 28. Pharm Res. 2010. PMID: 19862606
-
Multiple catalytically active thioredoxin folds: a winning strategy for many functions.Cell Mol Life Sci. 2010 Nov;67(22):3797-814. doi: 10.1007/s00018-010-0449-9. Epub 2010 Jul 13. Cell Mol Life Sci. 2010. PMID: 20625793 Free PMC article. Review.
-
Peroxiredoxin 1 and its role in cell signaling.Cell Cycle. 2009 Dec 15;8(24):4072-8. doi: 10.4161/cc.8.24.10242. Epub 2009 Dec 5. Cell Cycle. 2009. PMID: 19923889 Free PMC article. Review.
-
Native state fluctuations in a peroxiredoxin active site match motions needed for catalysis.Structure. 2022 Feb 3;30(2):278-288.e3. doi: 10.1016/j.str.2021.10.001. Epub 2021 Oct 21. Structure. 2022. PMID: 34678159 Free PMC article.
References
-
- Jacobson FS, Morgan RW, Christman MF, Ames BN. An alkyl hydroperoxide reductase from Salmonella typhimurium involved in the defense of DNA against oxidative damage. Purification and properties. J Biol Chem. 1989;264:1488–1496. - PubMed
-
- Niimura Y, Poole LB, Massey V. Amphibacillus xylanus NADH oxidase and Salmonella typhimurium alkyl-hydroperoxide reductase flavoprotein components show extremely high scavenging activity for both alkyl hydroperoxide and hydrogen peroxide in the presence of S. typhimurium alkyl-hydroperoxide reductase 22-kDa protein component. J Biol Chem. 1995;270:25645–25650. - PubMed
-
- Poole LB, Ellis HR. Flavin-dependent alkyl hydroperoxide reductase from Salmonella typhimurium. 1. Purification and enzymatic activities of overexpressed AhpF and AhpC proteins. Biochemistry. 1996;35:56–64. - PubMed
-
- Poole LB. Bacterial defenses against oxidants: mechanistic features of cysteine-based peroxidases and their flavoprotein reductases. Arch Biochem Biophys. 2005;433:240–254. - PubMed
-
- Wood ZA, Poole LB, Karplus PA. Structure of intact AhpF reveals a mirrored thioredoxin-like active site and implies large domain rotations during catalysis. Biochemistry. 2001;40:3900–3911. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

