3'UTR-located ALU elements: donors of potential miRNA target sites and mediators of network miRNA-based regulatory interactions
- PMID: 19455205
- PMCID: PMC2674674
3'UTR-located ALU elements: donors of potential miRNA target sites and mediators of network miRNA-based regulatory interactions
Abstract
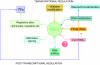
Recent research data reveal complex, network-based interactions between mobile elements and regulatory systems of eukaryotic cells. In this article, we focus on regulatory interactions between Alu elements and micro RNAs (miRNAs). Our results show that the majority of the Alu sequences inserted in 3'UTRs of analyzed human genes carry strong potential target sites for at least 53 different miRNAs. Thus, 3'UTR-located Alu elements may play the role of mobile regulatory modules that supply binding sites for miRNA regulation. Their abundance and ability to distribute a set of certain miRNA target sites may have an important role in establishment, extension, network organization, and, as we suppose - in the regulation and environment-dependent activation/inactivation of some elements of the miRNA regulatory system, as well as for a larger scale RNA-based regulatory interactions. The Alu-miRNA connection may be crucial especially for the primate/human evolution.
Keywords: retroelements; Alu elements; eukaryotic regulatory networks; evolution; miRNAs.
Figures


Similar articles
-
Novel Role of 3'UTR-Embedded Alu Elements as Facilitators of Processed Pseudogene Genesis and Host Gene Capture by Viral Genomes.PLoS One. 2016 Dec 29;11(12):e0169196. doi: 10.1371/journal.pone.0169196. eCollection 2016. PLoS One. 2016. PMID: 28033411 Free PMC article.
-
Multiple Alu Exonization in 3'UTR of a Primate-Specific Isoform of CYP20A1 Creates a Potential miRNA Sponge.Genome Biol Evol. 2021 Jan 7;13(1):evaa233. doi: 10.1093/gbe/evaa233. Genome Biol Evol. 2021. PMID: 33434274 Free PMC article.
-
Dynamic Variations of 3'UTR Length Reprogram the mRNA Regulatory Landscape.Biomedicines. 2021 Oct 28;9(11):1560. doi: 10.3390/biomedicines9111560. Biomedicines. 2021. PMID: 34829789 Free PMC article. Review.
-
The impact of miRNA target sites in coding sequences and in 3'UTRs.PLoS One. 2011 Mar 22;6(3):e18067. doi: 10.1371/journal.pone.0018067. PLoS One. 2011. PMID: 21445367 Free PMC article.
-
microRNAs and Alu elements in the p53-Mdm2-Mdm4 regulatory network.J Mol Cell Biol. 2014 Jun;6(3):192-7. doi: 10.1093/jmcb/mju020. J Mol Cell Biol. 2014. PMID: 24868102 Free PMC article. Review.
Cited by
-
Nuclear function of Alus.Nucleus. 2014 Mar-Apr;5(2):131-7. doi: 10.4161/nucl.28005. Epub 2014 Feb 4. Nucleus. 2014. PMID: 24637839 Free PMC article. Review.
-
MHC Class I Regulation: The Origin Perspective.Cancers (Basel). 2020 May 4;12(5):1155. doi: 10.3390/cancers12051155. Cancers (Basel). 2020. PMID: 32375397 Free PMC article.
-
Transcriptome-wide expansion of non-coding regulatory switches: evidence from co-occurrence of Alu exonization, antisense and editing.Nucleic Acids Res. 2013 Feb 1;41(4):2121-37. doi: 10.1093/nar/gks1457. Epub 2013 Jan 8. Nucleic Acids Res. 2013. PMID: 23303787 Free PMC article.
-
Alu RNA and their roles in human disease states.RNA Biol. 2021 Nov 12;18(sup2):574-585. doi: 10.1080/15476286.2021.1989201. Epub 2021 Oct 21. RNA Biol. 2021. PMID: 34672903 Free PMC article.
-
The DNA Habitat and its RNA Inhabitants: At the Dawn of RNA Sociology.Genomics Insights. 2013 Mar 4;6:1-12. doi: 10.4137/GEI.S11490. eCollection 2013. Genomics Insights. 2013. PMID: 26217106 Free PMC article.
References
-
- Mighell AJ, Markham AF, Robinson PA. Alu sequences. FEBS Letters. 417(1):1–5. - PubMed
-
- Batzer Mark A, Deininger Prescott L. Alu repeats and human genomic diversity. Nature Reviews Genetics. 2002 May;3 - PubMed
-
- Neil R Smalheiser, Vetle I. Torvik Mammalian microRNAs derived from genomic repeats. Trends in Genetics. 2005 Jun;21(6) - PubMed
LinkOut - more resources
Full Text Sources
