The Influenza Primer Design Resource: a new tool for translating influenza sequence data into effective diagnostics
- PMID: 19453490
- PMCID: PMC4634328
- DOI: 10.1111/j.1750-2659.2007.00031.x
The Influenza Primer Design Resource: a new tool for translating influenza sequence data into effective diagnostics
Abstract
Background: Recent outbreaks of highly pathogenic avian influenza and multiple occurrences of zoonotic infection and deaths in humans have sparked a dramatic increase in influenza research. In order to rapidly identify and help prevent future influenza outbreaks, numerous laboratories around the world are working to develop new nucleotide-based diagnostics for identifying and subtyping influenza viruses. While there are several databases that have been developed for manipulating the vast amount of influenza genetic data that have been produced, significant progress can still be made in developing tools for translating the genetic data into effective diagnostics.
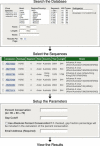
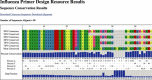
Description: The Influenza Primer Design Resource (IPDR) is the combination of a comprehensive database of influenza nucleotide sequences and a web interface that provides several important tools that aid in the development of oligonucleotides that may be used to develop better diagnostics. IPDR's database can be searched using a variety of criteria, allowing the user to align the subset of influenza sequences that they are interested in. In addition, IPDR reports a consensus sequence for the alignment along with sequence polymorphism information, a summary of most published primers and probes that match the consensus sequence, and a Primer3 analysis of potential primers and probes that could be used for amplifying the sequence subset.
Conclusions: The IPDR is a unique combination of bioinformatics tools that will greatly aid researchers in translating influenza genetic data into diagnostics, which can effectively identify and subtype influenza strains. The website is freely available at http://www.ipdr.mcw.edu.
Figures


Similar articles
-
Influenza research database: an integrated bioinformatics resource for influenza research and surveillance.Influenza Other Respir Viruses. 2012 Nov;6(6):404-16. doi: 10.1111/j.1750-2659.2011.00331.x. Epub 2012 Jan 20. Influenza Other Respir Viruses. 2012. PMID: 22260278 Free PMC article.
-
FAO-OIE-WHO Joint Technical Consultation on Avian Influenza at the Human-Animal Interface.Influenza Other Respir Viruses. 2010 May;4 Suppl 1(Suppl 1):1-29. doi: 10.1111/j.1750-2659.2009.00114.x. Influenza Other Respir Viruses. 2010. PMID: 20491978 Free PMC article.
-
RExPrimer: an integrated primer designing tool increases PCR effectiveness by avoiding 3' SNP-in-primer and mis-priming from structural variation.BMC Genomics. 2009 Dec 3;10 Suppl 3(Suppl 3):S4. doi: 10.1186/1471-2164-10-S3-S4. BMC Genomics. 2009. PMID: 19958502 Free PMC article.
-
Website for avian flu information and bioinformatics.Sci China C Life Sci. 2009 May;52(5):470-3. doi: 10.1007/s11427-009-0065-9. Epub 2009 May 27. Sci China C Life Sci. 2009. PMID: 19471870 Review.
-
RT-PCR/electrospray ionization mass spectrometry approach in detection and characterization of influenza viruses.Expert Rev Mol Diagn. 2011 Jan;11(1):41-52. doi: 10.1586/erm.10.107. Expert Rev Mol Diagn. 2011. PMID: 21171920 Review.
Cited by
-
Multiplex assay for simultaneously typing and subtyping influenza viruses by use of an electronic microarray.J Clin Microbiol. 2009 Feb;47(2):390-6. doi: 10.1128/JCM.01807-08. Epub 2008 Dec 10. J Clin Microbiol. 2009. PMID: 19073867 Free PMC article.
-
Rapid semiautomated subtyping of influenza virus species during the 2009 swine origin influenza A H1N1 virus epidemic in Milwaukee, Wisconsin.J Clin Microbiol. 2009 Sep;47(9):2779-86. doi: 10.1128/JCM.00999-09. Epub 2009 Jul 29. J Clin Microbiol. 2009. PMID: 19641066 Free PMC article.
-
Development of a rapid automated influenza A, influenza B, and respiratory syncytial virus A/B multiplex real-time RT-PCR assay and its use during the 2009 H1N1 swine-origin influenza virus epidemic in Milwaukee, Wisconsin.J Mol Diagn. 2010 Jan;12(1):74-81. doi: 10.2353/jmoldx.2010.090095. Epub 2009 Dec 3. J Mol Diagn. 2010. PMID: 19959800 Free PMC article.
-
Development and clinical testing of a simple, low-density gel element array for influenza identification, subtyping, and H275Y detection.J Virol Methods. 2014 Nov;208:152-9. doi: 10.1016/j.jviromet.2014.07.019. Epub 2014 Jul 24. J Virol Methods. 2014. PMID: 25066276 Free PMC article.
-
Rapid multiplex reverse transcription-PCR typing of influenza A and B virus, and subtyping of influenza A virus into H1, 2, 3, 5, 7, 9, N1 (human), N1 (animal), N2, and N7, including typing of novel swine origin influenza A (H1N1) virus, during the 2009 outbreak in Milwaukee, Wisconsin.J Clin Microbiol. 2009 Sep;47(9):2772-8. doi: 10.1128/JCM.00998-09. Epub 2009 Jul 29. J Clin Microbiol. 2009. PMID: 19641063 Free PMC article.
References
-
- Webster RG, Govorkova EA. H5N1 influenza – continuing evolution and spread. N Engl J Med 2006; 355:2174–2177. - PubMed
-
- Hampson AW, Mackenzie JS. The influenza viruses. Med J Aust 2006; 10(Suppl.):S39–S43. - PubMed
-
- Macken C, Lu H, Goodman J, Boykin L. The value of a database in surveillance and vaccine selection; in Osterhaus ADME, Cox N, Hampson AW. (ed): Options for the Control of Influenza IV. Amsterdam: Elsevier Science, 2001.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

